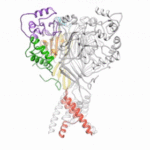
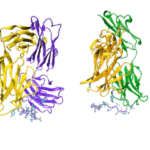
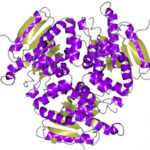
Using protein crystallography at the Advanced Light Source (ALS), investigators at Oregon Health and Science University visualized an acid-sensing ion channel, ASIC1a, in it’s elusive closed state for the first time. An important cell membrane protein in the central nervous system, ASIC1a plays a role in sensing pain and in forming memories of fear. The researchers used protein crystals of ASIC1a channels purified in the closed state and compiled X-ray data from ALS Beamline 5.0.2 and the Advanced Photon Source at Argonne National Lab to build a model of the closed channel structure. Defining the closed structure enabled the researchers to generate a comprehensive molecular model of how ASIC1a toggles between the closed, open, and desensitized states.
Read more from the ALS.