Human skin is teeming with microbial life, and not just bacteria. An international collaboration among researchers at Berkeley Lab, led by Hoi-Ying Holman, the Medical University of Graz in Austria and at the University of Regensburg in Germany, found representatives of the domain archea–which inhabit some of the most extreme environments on the planet–coexisting alongside more run-of-the-mill microbes. What’s more, according to the paper published in Scientific Reports, the relative abundance of archea varies with a person’s age. Holman, a senior author on the report, is head of Chemical Ecology in the Earth & Environmental Sciences Area and has a secondary affiliation in Biosciences in the Molecular Biophysics and Integrated Bioimaging (MBIB) division. Using the Advanced Light Source (ALS), her group developed a rapid and label-free method to screen cells and tell if they’re bacteria or archaea. Read more from the Berkeley Lab News Center.
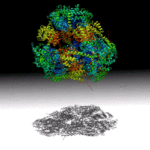
An Atomic-Level Snapshot of Bacterial Microcompartment Architecture
Researchers led by Berkeley Lab Biosciences’ Cheryl Kerfeld (MBIB), who holds a joint appointment at the MSU-DOE Plant Research Laboratory, have provided the first atomic-level resolution picture of an intact bacterial microcompartment (BMC) shell. The intact shell and component proteins from the saltwater bacterium Haliangium ochraceum were crystallized at Berkeley Lab; X-ray diffraction data were collected at the Lab’s Advanced Light Source (ALS) and at the Stanford Synchrotron Radiation Lightsource. Details of the BMC shell’s structure and composition were published in the journal Science.
A Surprise Just Beneath the Surface in Carbon Dioxide Experiment
Research co-led by Berkeley Lab researchers Junko Yano (MBIB) and Ethan Crumlin at the Advanced Light Source (ALS), with collaborators at Caltech’s Joint Center for Artificial Photosynthesis (JCAP), has revealed a surprising driver of a chemical process to reformulate carbon dioxide into more useful compounds. X-ray experiments coupled with theoretical models showed that oxygen atoms near the surface of a copper sample had a more dramatic effect on the early stages of a reaction with carbon dioxide than earlier theories could account for. This work could help make reactions more efficient in converting carbon dioxide into liquid fuels and other products. Read more in the News Center release.
A Hollow Pyramid Unlocks Principles of Protein Architecture
Researchers at UCLA have designed a hollow, pyramid-shaped protein with a controllable cavity size that may aid the capture and release of smaller compounds. The tools used in this work, including small angle X-ray scattering techniques at the SIBYLS beamline in the Advanced Light Source (ALS), will help analyze and optimize designed-protein assemblies and understand their behavior in solution. Read more in the ALS Science Brief.
Designing Cyclic Oligomers: Greater than the Sum of Their Parts
Cyclic proteins that assemble from multiple identical subunits (homo-oligomers) play key roles in many biological processes, including enzymatic catalysis and function and cell signaling. Researchers in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division worked with University of Washington’s David Baker, who led a team to design in silico and crystallize self-assembling cyclic homo-oligomer proteins.
Was this page useful?