As a young man, Kenneth Sauer joined Berkeley Lab four years after arriving in Berkeley for his postdoctoral position with famed chemist Melvin Calvin. By that time, he had accepted an assistant professorship in UC Berkeley’s Department of Chemistry to continue what would be his life’s scientific work on the intricate physical process of photosynthesis. He remained active for over 50 years and was, most recently, a professor emeritus of chemistry at UC Berkeley. Sauer died at the age of 91 following a brief illness on November 6, 2022.
Researchers Capture Elusive Missing Step in Photosynthesis
After decades of effort, scientists have revealed atomic-scale details of the water splitting step of photosynthesis, the chemical process that generates the air we breathe. The latest work adds to our understanding of photosynthesis and will aid the development of fully renewable alternative energy sources.
Chloro-phylling in the Answers to Big Questions
A team of scientists, including many in the Molecular Biophysics and Integrated Bioimaging Division, uncovered new details about the reaction that powers photosynthesis. Understanding this reaction could lead to world-changing advances in technology, medicine, or energy––and also gives insight into how the enzyme photosystem II produces the oxygen we breathe. Their latest work was recently published in Nature Communications and two of the authors, Vittal Yachandra and Philipp Simon, spoke with Strategic Communications about that, shooting stuff with lasers, and why they chose this field of research.
New Technique Gets the Drop On Enzyme Reactions
As part of an international collaboration, researchers at Lawrence Berkeley National Laboratory (Berkeley Lab), the Diamond Light Source synchrotron facility, and Oxford and Bristol Universities in England have developed a novel sample delivery system that expands the limited toolkit for performing dynamic structural biology studies of enzyme catalysis, which have so far mostly been limited to a small number of light-driven enzymes.
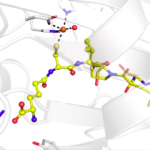
Enzyme Action Movie Shows How Nature Makes Penicillins
Scientists who specialize in studying the atom-by-atom choreography of enzymes have revealed new insights into the function of isopenicillin N synthase, an enzyme needed to produce some of the world’s most critical antibiotics.
Was this page useful?