After decades of effort, scientists have finally glimpsed a key step in the process by which nature creates the oxygen we breathe.
Researchers from Berkeley Lab – led by senior scientists Junko Yano and Vittal Yachandra, and staff scientist Jan Kern in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division – SLAC National Accelerator Laboratory, and collaborators other institutions, have finally succeeded in laying bare a key secret of photosynthesis. Using SLAC’s Linac Coherent Light Source (LCLS) and the SPring-8 Angstrom Compact free electron LAser (SACLA) in Japan, they captured in atomic detail what transpires in the final moments leading up to the release of oxygen.
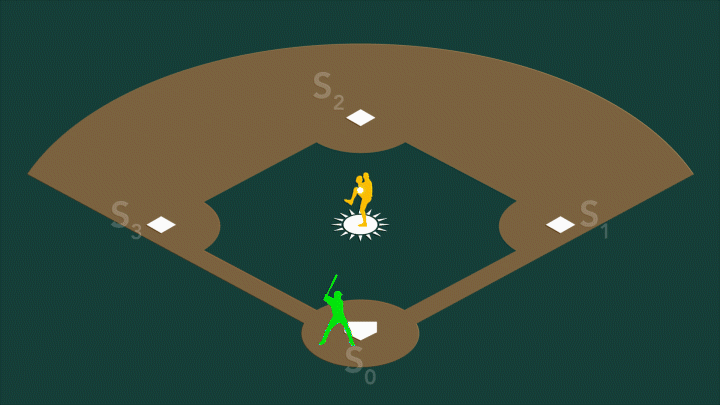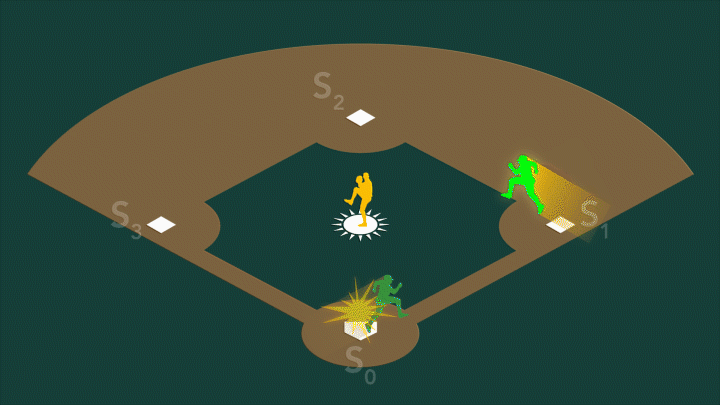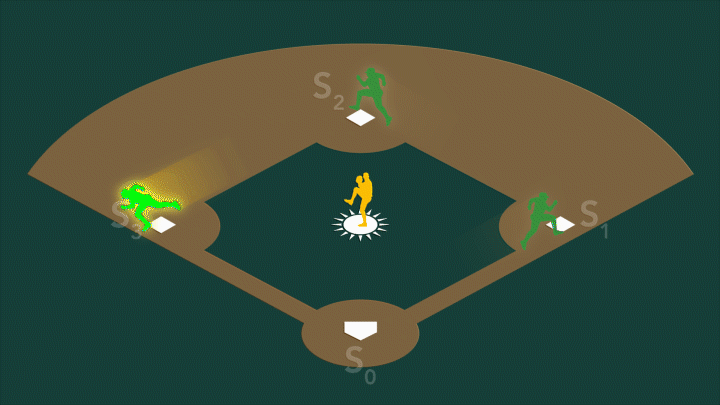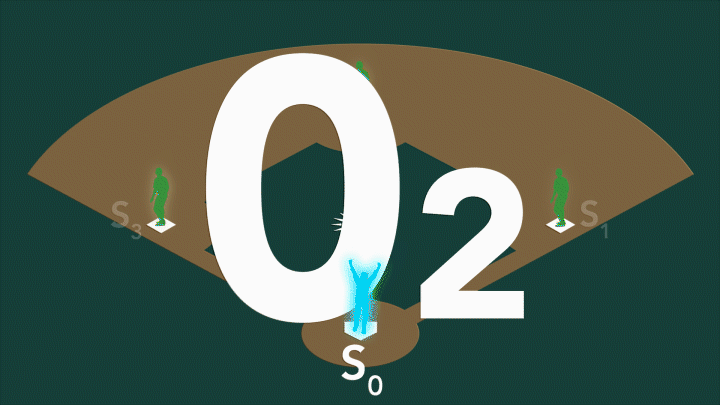
During photosynthesis, a protein complex called photosystem II found in plants, algae, and cyanobacteria harvests sunlight and uses it to split water, producing the oxygen we breathe. Photosystem II’s oxygen-evolving center – a cluster of four manganese atoms and one calcium atom connected by oxygen atoms – facilitates a series of challenging chemical reactions that accomplish this feat. When exposed to sunlight, the center cycles through four stable oxidation states, known as S0 through S3.
The researchers examined this center by exciting tiny samples of photosynthetic molecules from cyanobacteria with optical light. They then probed the molecules with ultrafast X-ray pulses from LCLS and SACLA, using a bespoke conveyor belt-inspired instrument designed Isabel Bogacz, a graduate student research assistant, and Philipp Simon, a postdoctoral researcher, in the Yano/Yachandra/Kern group.
Using this technique, the scientists for the first time imaged the transient S4 state, where two atoms of oxygen bond together and an oxygen molecule is released. The data, reported in the journal Nature, showed that there are additional steps in this reaction that had never been seen before.
After gathering all the data – taken over six years – the team had to analyze it and piece together structural maps of the molecules as they change during the reaction. This work was made possible by special software for data merging developed by senior scientist Nicholas Sauter and research scientist Aaron Brewster, and by programs for structure determination developed by Paul Adams, Associate Laboratory Director for Biosciences. The data analysis was performed by project scientist Asmit Bhowmick, Rana Hussein of Humboldt University, Bogacz, and Simon.
Additional MBIB researchers who contributed to this work include: Ruchira Chatterjee, Margaret Doyle, Medhanjali Dasgupta, James Holton, Corey Kaminsky, Stephen Keable, In-Sik Kim, Hiroki Makita, Nigel Moriarty, Isabela Nangca, Daniel Paley, and Miao Zhang.




