Naturally occurring proteins—chains of amino acids that fold into functional, three-dimensional shapes—are believed to represent just a small fraction of the universe of all possible permutations of amino-acid sequences and folds.  How can we begin to systematically sift through those permutations to find and engineer from scratch (de novo) proteins with the characteristics desired for medical, environmental, and industrial purposes? To address this question, a team led by researchers from the Institute for Protein Design at the University of Washington have published a landmark study that used both protein crystallography (Beamlines 8.2.1 in the Berkeley Center for Structural Biology and 8.3.1) and small-angle x-ray scattering (SAXS; SIBYLS Beamline) at the ALS to validate the computationally designed structures of novel proteins with repeated motifs. The results show that the protein-folding universe is far larger than realized, opening up a wide array of new possibilities for biomolecular engineering. Read the ALS Science Highlight.
How can we begin to systematically sift through those permutations to find and engineer from scratch (de novo) proteins with the characteristics desired for medical, environmental, and industrial purposes? To address this question, a team led by researchers from the Institute for Protein Design at the University of Washington have published a landmark study that used both protein crystallography (Beamlines 8.2.1 in the Berkeley Center for Structural Biology and 8.3.1) and small-angle x-ray scattering (SAXS; SIBYLS Beamline) at the ALS to validate the computationally designed structures of novel proteins with repeated motifs. The results show that the protein-folding universe is far larger than realized, opening up a wide array of new possibilities for biomolecular engineering. Read the ALS Science Highlight.
X-ray Studies at SLAC and Berkeley Lab Aid Search for Ebola Cure
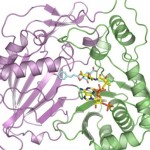
Recently, scientists from University of California, San Francisco, performed research at two national laboratories to determine protein structures that c0uld be the key to preventing Ebola infection. Alexander Kintzer and Robert Stroud, used two structural biology beamlines (5.0.2 and 8.3.1) at Berkeley Lab’s Advanced Light Source, to determine in atomic detail how a potential drug molecule fits into and blocks a channel in cell membranes that Ebola and related “filoviruses” need to infect victims’ cells. The study, published March 9 in Nature, marks an important step toward finding a cure for Ebola and other diseases that depend on the channel. Read more at the SLAC News Center.
New Way to Reduce Plant Lignin Could Lead to Cheaper Biofuels
Aymerick Eudes and Dominique Loqué of the Joint Bioenergy Institute (JBEI) led a study that shows for the first time that an enzyme can be tweaked to reduce lignin in plants. Their technique could help lower the cost of converting biomass into carbon-neutral fuels to power your car and other sustainably developed bio-products. The crystal structure of this enzyme was solved using data collected in the Berkeley Center for Structural Biology at the Advanced Light Source. Read more on the Berkeley Lab News Center.
Improving Meningococcal Vaccines
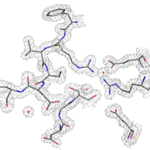
Two recently licensed vaccines against bacterial meningitis contain a bacterial surface protein antigen known as Factor H binding protein (FHbp). The native form of this protein can have low thermal stability, which limits its potential use as an antigen in vaccines. After engineering a more stable Factor H binding protein antigen, scientists from UC San Francisco Benioff Children’s Hospital Oakland determined the structure of the stabilized vaccine with the help of protein crystallography at the Advanced Light Source (ALS) in the Berkeley Center for Structural Biology (Beamline 5.0.1). Read more in the ALS Science Brief.
CRISPR/Cas9: Ready for Action
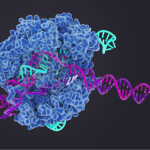
The CRISPR/Cas9 bacterial genomic editing system identifies and cleaves complementary target sequences in foreign DNA. CRISPR (clustered regularly interspaced short palindromic repeats)–associated (Cas) protein Cas9 begins its work by RNA-guided DNA unwinding to form an RNA-DNA hybrid and displacing a DNA strand inside the protein. Upon binding, Cas9 reorganizes into an R-loop complex that is necessary for it to perform its function. A recent article published in Science describes work done to uncover the structural basis of Cas9’s function.
Was this page useful?