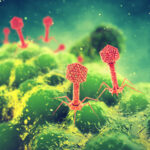
As conventional antibiotics continue to lose effectiveness against evolving pathogens, scientists are keen to employ the bacteria-killing techniques perfected by bacteriophages (phages), the viruses that infect bacteria. One major challenge is the difficulty of studying individual phage proteins and determining precisely how the virus wields these tools to kill their host bacteria. A team of researchers from Berkeley Lab, UC Berkeley, and Texas A&M University worked together on a high-throughput genetic screen to identify which part of the bacteria the phages were targeting.
Ten Simple Rules for Getting and Giving Credit for Data
Generating and analyzing data associated with scientific research can be challenging and complicated, to say the least. But the importance of sharing and giving credit to those who produced the data is foundational to furthering the impact of the work. Learn more about ten simple rules for getting and giving credit for data.
Small-scale Changes in Environment Can Have Large Effects on Microbial Communities
A Berkeley Lab team analyzed the genotypes and phenotypes of several Arthrobacter strains to correlate cellular functions to their location at varying depths within a single sediment core and in nearby groundwater. They found that Arthrobacter, as a genus, has remarkable flexibility in altering its suites of carbon degradation genes. This genomic variation was found to be linked to the individual strain’s environment and is the basis for Arthrobacter’s ability to break down a wide variety of complex carbon sources.
All-star Scientific Team Seeks to Edit Entire Microbiomes with CRISPR
CRISPR enzymes are like super scissors: they cut, delete, and add genes to a specific kind of cell, one at a time. But now, UC Berkeley faculty and Biosciences Area researchers have figured out how to add or modify genes within a microbial community of many different species, coining the phrase, “community editing.”
Project Jupyter: Computer Code that Transformed Science
Computer code co-developed by a scientist from Lawrence Berkeley National Laboratory (Berkeley Lab) and embraced by the global science community over two decades has been hailed by Nature as one of “ten computer codes that transformed science.”
- 1
- 2
- 3
- 4
- Next Page »
Was this page useful?