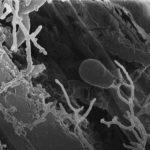
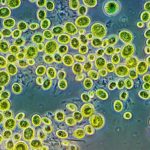
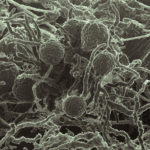
Within a forest’s soil, a microbiome of bacteria, viruses and fungi process carbon and nitrogen, paving the way for future plants and trees to grow. However, fire changes the microbes within the soil. Recently, JGI collaborators worked to understand which microbes in the soil persist after a wildfire — and why they thrive. Their results appear in Nature Microbiology.
JGI Helps Uncover Novel Chemicals from an Unexpected Source: Gut Fungi
Anaerobic fungi, which die in the presence of oxygen, thrive in herbivore guts and help them digest their host’s last leafy meal. In their evolutionary history, these fungi branched off early from aerobic fungi, which can breathe oxygen — just like we do. Oxygen is a rich source of energy, and because anaerobic fungi can’t harness it, scientists long held that these fungi don’t have the energy to make complex compounds called natural products. Yet, analyzing the genomes and genome products of four anaerobic fungal species has revealed that this group is unexpectedly powerful: they can whip up dozens of complex natural products, including new ones. The work was partly enabled by the “Facilities Integrating Collaborations for User Science” (FICUS) collaborative science initiative between the JGI and the Environmental Molecular Sciences Laboratory. Read the full science highlight on the JGI website.
JGI Launches Data Portal for Algae
A JGI team led by Algal Genomics Program lead Igor Grigoriev and data scientist Alan Kuo have unveiled PhycoCosm in the Nucleic Acids Research journal. The genome portal reinforces the JGI’s new strategic focus on exploring algal biology, diversity, and ecology. Read more here on the JGI website.
Fungal Enzyme Clusters Promote Efficient Biomass Breakdown
Fungi, particularly those found in the digestive tracts of ruminant herbivores such as cattle, sheep, and goats, are very good at overcoming the resistance of plant cell walls to degradation—a major hurdle in the quest to produce sustainable fuels and chemicals from bioenergy feedstocks. Now, an international group of researchers has identified protein scaffolds in anaerobic gut fungi that provide docking sites for various enzymes, keeping them in place so that they can work together more effectively. As reported in the May 26 issue of Nature Microbiology, the structures are analogous to cellulosome complexes in anerobic bacteria, but this is the first time they have been found in fungi.
DOE JGI Finds A New Major Gene Expression Regulator in Fungi
Just four letters – A, C, T, and G – make up an organism’s genetic code. Changing a single letter, or base, can lead to changes in protein structures and functions, impacting an organism’s traits. In addition, though, subtler changes can and do happen, involving modifications of the DNA bases themselves. In the May 8, 2017 issue of Nature Genetics, a team led by scientists at the U.S. Department of Energy Joint Genome Institute (JGI), a DOE Office of Science User Facility, report the prevalence of modifications in the earliest branches of the fungal kingdom. Read more in the JGI press release.
Was this page useful?