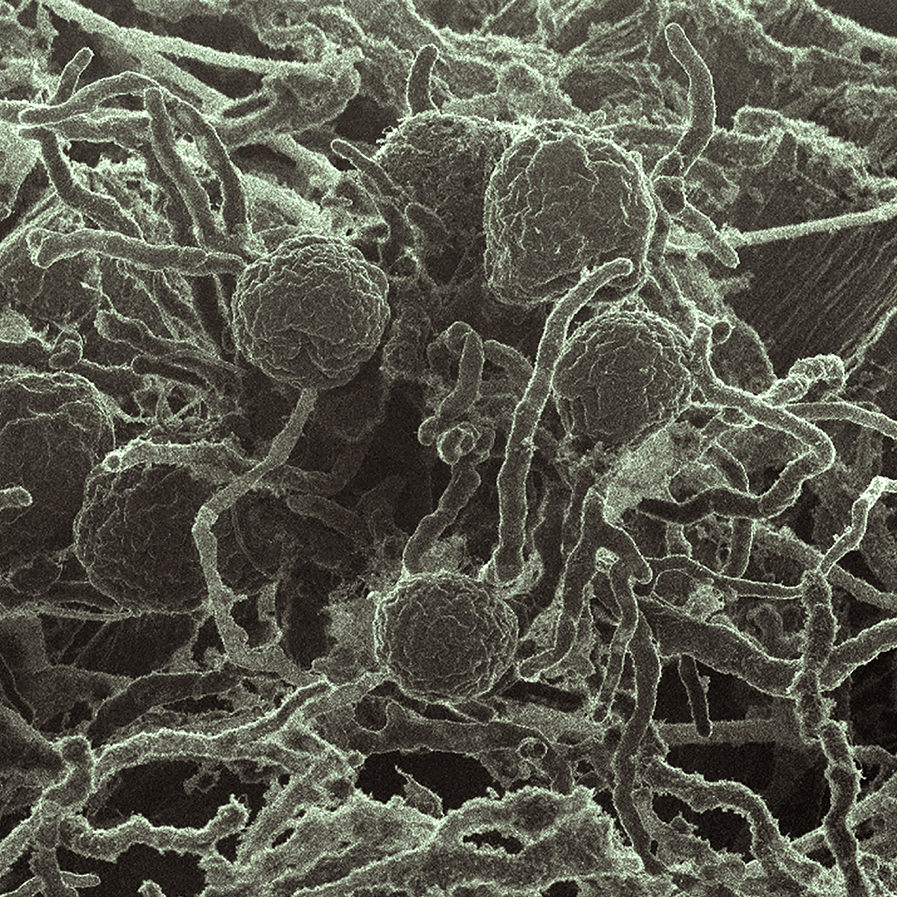
 Fungi, particularly those found in the digestive tracts of ruminant herbivores such as cattle, sheep, and goats, are very good at overcoming the resistance of plant cell walls to degradation—a major hurdle in the quest to produce sustainable fuels and chemicals from bioenergy feedstocks.
Fungi, particularly those found in the digestive tracts of ruminant herbivores such as cattle, sheep, and goats, are very good at overcoming the resistance of plant cell walls to degradation—a major hurdle in the quest to produce sustainable fuels and chemicals from bioenergy feedstocks.
Now, an international group of researchers has identified protein scaffolds in anaerobic gut fungi that provide docking sites for various enzymes, keeping them in place so that they can work together more effectively. As reported in the May 26 issue of Nature Microbiology, the structures are analogous to cellulosome complexes in anerobic bacteria, but this is the first time they have been found in fungi. The work harnessed the capabilities of two Department of Energy (DOE) Office of Science User Facilities: the DOE Joint Genome Institute (JGI) at Berkeley Lab and the Environmental Molecular Sciences Laboratory (EMSL) at Pacific Northwest National Laboratory (PNNL).
A team at UC Santa Barbara, led by assistant professor of bioengineering Michelle O’Malley, a co-senior author on the paper, collected animal fecal samples and isolated anerobic fungi belonging to the Neocallimastigomycota family, which has not, to date, been well-characterized. Three of the isolated fungi were sequenced and annotated by a team at DOE JGI which included Alan Kuo, Stephen Mondo, Asaf Salamov, Kurt LaButti, Zhiying Zhao, Jennifer Chiniquy, Kerrie Barry, and Igor Grigoriev.
“The three genomes are really well resolved to the point where you can start looking at what’s there, what’s regulating enzyme production, and how enzymes have evolved,” O’Malley said.
Igor Grigoriev, who heads the DOE JGI’s Fungal Genomics Program and was co-senior author on the paper, noted that the Facilities Integrating Collaborations for User Science (FICUS) initiative, established by JGI and EMSL in 2014, enabled the multi-omics approach that was crucial to the research. “Though the fungal cellulosome was discovered through proteomics, we needed genomics and transcriptomics to decode all its parts,” he said. A detailed understanding of how these fungi are able to efficiently degrade plant material provides a basis for harnessing those capabilities through synthetic biology or metabolic engineering. Read more on the DOE JGI website.