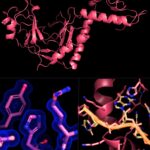
MBIB researchers have conducted the first ever structural analysis of a key protein involved in DNA damage repair and cancer. While the prevailing narrative surrounding cancer chemotherapy has historically focused on DNA damage repair pathways, findings from this study underscore the key role of RNA-mediated processes on chemotherapy response, which could have major implications for cancer treatment outcomes.
Researchers Leverage SAXS to Understand Aspect of Microbial Metabolism
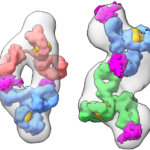
A team of Molecular Biophysics and Integrated Bioimaging Division researchers used synchrotron technology unique to the Advanced Light Source (ALS) at Berkeley Lab to probe the conformational states behind electron bifurcation.
How X-rays Could Make Reliable, Rapid COVID-19 Tests a Reality
An imaging technique pioneered by Berkeley Lab is helping reveal the best antibodies to test for in rapid and reliable COVID-19 detection. Although current tests such as polymerase chain reaction (PCR) are highly accurate, these samples must be sent to an accredited lab for testing, causing a longer wait time for results. Michal Hammel, a research scientist in the Molecular Biophysics and Integrated Bioimaging Division, and Curtis D. Hodge led a study that could help get reliable, self-administered tests with instant results on the market.
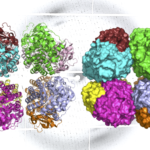
Study Finds ‘Missing Link’ in the Evolutionary History of Carbon-Fixing Protein Rubisco
In a study appearing in Nature Plants, researchers from UC Davis, UC Berkeley, and Berkeley Lab report the discovery and characterization of a previously undescribed lineage of form I rubisco – one that the researchers suspect diverged from form I rubisco prior to the evolution of cyanobacteria. The novel lineage, called form I’ rubisco, gives researchers new insights into the structural evolution of form I rubisco, potentially providing clues as to how this enzyme changed the planet.
The work was led by Patrick Shih, a UC Davis assistant professor and the director of Plant Biosystems Design at the Joint BioEnergy Institute (JBEI), and Doug Banda, a postdoctoral scholar in his lab.
SIBYLS Sheds Light on Enzyme that Regulates Blood-clotting Factor
A group of researchers at Washington University School of Medicine have used the capabilities available at the Advanced Light Source’s SIBYLS beamline to gain insight into an enzyme that functions in blood clotting. ADAMTS13, which stands for a disintegrin and metalloproteinase with thrombospondin-1 repeats, member 13, is a multi-domain protease enzyme whose catalytic mechanism involves a metal. It is the only known protein to regulate the adhesive function of von Willebrand factor (VWF), a blood clotting protein involved in hemostasis.
Was this page useful?