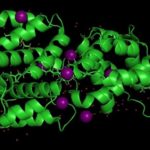
The Helical Carotenoid Protein 2 (HCP2) protein is an ancestor of proteins that are known to protect against damage caused by excess light exposure. Researchers in the laboratory of Cheryl Kerfeld, guest faculty in the Environmental Genomics & Systems Biology (EGSB) Division, are the first to structurally and biophysically analyze a protein from the HCP family. This HCP protein family was discovered recently by Kerfeld and the members of her lab, who are based in EGSB and at Michigan State University (MSU). To solve the molecular structure of HCP2, X-ray diffraction was measured at beam line 5.0.2 in the Berkeley Center for Structural Biology of the Advanced Light Source (ALS). The structure was refined using Phenix, a software suite for automated determination of molecular structures developed under the direction of Paul Adams, Molecular Biophysics and Integrated Bioimaging Division Director. Read more in the MSU-DOE Plant Research Laboratory news story.
Revealing the Shapes of Molecular Machines
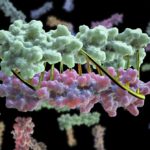
Within each cell of the human body, thousands of molecular machines are at work. They transport nutrients and biochemicals into and out of our cells, build other tiny machines, and even move our cells around. To understand how these molecular machines work, scientists create three-dimensional pictures using electron cryomicroscopy (cryo-EM), catching these machines in different shapes that give insight into their function. Now researchers at Berkeley Lab and their international collaborators who write and distribute the Phenix software suite have developed a new set of computational tools for automated structure determination from cryo-EM data.
Researchers Identify Openings for Shuttering Virus Factories
A team led by Mary Estes of the Baylor College of Medicine used rotavirus as a model to study some of the proteins involved in making the cytoplasmic compartments in which many DNA and RNA virus pathogens replicate. Banumathi Sankaran, a research scientist in the Berkeley Center for Structural Biology (BSCB) at the Advanced Light Source, collected the X-ray data at the BCSB Beamline 5.0.1 that were used to solve the three-dimensional structures of nonstructural protein NSP2. Understanding the functions of proteins that make these compartments could offer an avenue for disrupting virus production. The team published their findings in Proceedings of the National Academy of Sciences.
Programming Proteins to Pair Perfectly
Bioscientists at the Advanced Light Source (ALS) at Berkeley Lab lent their expertise to a project led by scientists at the University of Washington to design proteins in the lab that zip together like DNA. The technique could enable the design of protein nanomachines to help diagnose and treat disease, allow for more precise engineering of cells, and perform a variety of other tasks.
NIH Awards $9.3M for Further Development of PHENIX Structural Biology Software
The National Institutes of Health (NIH) has awarded $9.3 million to Berkeley Lab to support ongoing development of PHENIX, a software suite for solving three-dimensional macromolecular structures. Officially launched in 2000, the project is a collaboration among researchers based at Berkeley Lab, Los Alamos National Laboratory, Cambridge University, and Duke University. “The impetus behind PHENIX is a desire to make the computational aspects of crystallography more automated, reducing human error and speeding solutions,” said PHENIX principal investigator Paul Adams, director of Berkeley Lab’s Molecular Biophysics and Integrated Bioimaging Division (MBIB). Read more from the Berkeley Lab News Center.
Was this page useful?