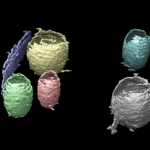
A team of Berkeley Lab scientists has developed a machine-learning pipeline to facilitate segmentation of tomograms of cell membrane structures. The project was an LDRD-funded collaboration among Chao Yang from the Applied Mathematics and Computational Research Division, and Nick Sauter and Karen Davies from the Molecular Biophysics and Integrated Bioimaging (MBIB) Division.
Machine Learning Tackles Long COVID
A new machine learning tool developed by a team of researchers led by Justin Reese of Berkeley Lab and Peter Robinson of Jackson Lab analyzes electronic health records to find symptoms in common between people who have been diagnosed with long COVID and to define subtypes of the condition.
Agile BioFoundry, Lygos to Advance Machine Learning in Organic Acid Development
The Agile BioFoundry and Lygos, Inc. are joining forces to generate the largest multi-omics dataset for guiding the development of organic acids. Over the course of the project, scientists will produce more than 500,000 data points from a series of experiments. ABF is now using its artificial neural networks to train machine learning algorithms and provide actionable recommendations to help optimize strain performance, increase operational efficiencies and enhance production.
Machine Learning Takes on Synthetic Biology: Algorithms Can Bioengineer Cells for You
Engineering biological systems to specification–for example, designing a microbe to produce a cancer-fighting agent–requires a detailed mechanistic understanding of how all the parts of a cell work. Typically, this knowledge is acquired through years of painstaking work and a fair amount of trial and error. But Berkeley Lab scientists have created an Automated Recommendation Tool (ART) that adapts machine learning algorithms to the needs of synthetic biology to guide development systematically. With a limited set of training data, the algorithms are able to predict how changes in a cell’s DNA or biochemistry will affect its behavior, then make recommendations for the next engineering cycle along with probabilistic predictions for attaining the desired goal. The work was led by Hector Garcia Martin, a researcher in Berkeley Lab’s Biological Systems and Engineering (BSE) Division and Tijana Radivojevic, a BSE data scientist. In a pair of papers recently published in the journal Nature Communications, they presented the algorithm and demonstrated its capabilities.
Read more in the Berkeley Lab News Center.
Using Machine Learning to Estimate COVID-19’s Seasonal Cycle
A cross-disciplinary team of Berkeley Lab scientists with expertise in climate modeling, data analytics, machine learning, and geospatial analytics is launching a project to determine if the novel coronavirus might be seasonal. The team will apply machine-learning methods to a plethora of health and environmental datasets, combined with high-resolution climate models and seasonal forecasts.
Was this page useful?