After 9-month national search, Nigel Mouncey, currently Research and Development Director for Bioengineering and Bioprocessing at Dow AgroSciences LLC, has been selected as the Director of the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility. Mouncey will be the DOE JGI’s fourth Director in its 20-year history. He succeeds Eddy Rubin, who announced his retirement in March 2016 after a dozen years as Director, and DOE JGI Strategic Planning Deputy Axel Visel, who has served as Interim Director since then. Read more about Mouncey’s selection on the DOE JGI website.
Engineering a More Efficient System for Harnessing Carbon Dioxide
Tapping the DNA synthesis expertise of the DOE Joint Genome Institute (JGI), a team from the Max-Planck-Institute (MPI) for Terrestrial Microbiology in Marburg, Germany has reverse engineered a biosynthetic pathway for more effective carbon fixation. Read more at JGI News.
DOE JGI Summer Intern Author on GOLD Article
Kaushal Sharma, one of five Antioch High School rising juniors who participated in internships at the DOE Joint Genome Institute (JGI) this summer through the Biotech Partners program is now an author on a Nucleic Acids Research publication. The paper describes the version 6 data updates and feature enhancements to the Genomes OnLine Database (GOLD) administered by JGI. T.B.K Reddy, Kaushal’s mentor, described his student’s contributions as reviewing close to 3,800 metagenomes to verify geographic locations and assign latitude and longitude values “This exercise helped us in filling or updating geographic information for nearly 1,100 samples,” said Reddy. Kaushal also reviewed and assigned NCBI taxonomy name and IDs for nearly 20,000 public/private mtagenome biosamples in GOLD. “Besides these, he curated several small sets of phylogeny, biosample metadata tasks we assigned.” What did you do for your summer vacation?
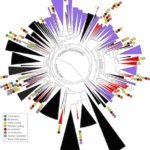
New Bacteria Groups, and Stunning Diversity, Discovered Underground
Lawrence Berkeley National Laboratory (Berkeley Lab) and UC Berkeley researchers have uncovered new clues about the roles of subsurface microbes in globally important cycles. Jill Banfield, senior faculty scientist in the Earth & Environmental Systems Area and professor at UC Berkeley, led the research team that studied soil and water samples containing subsurface microbes collected at a Colorado River basin field site. DNA sequencing of these microbes was performed at the Joint Genome Institute (JGI), a DOE Office of Science User Facility.
As reported online October 24 in the journal Nature Communications, the scientists netted genomes from 80 percent of all known bacterial phyla, a remarkable degree of biological diversity at one location. They also discovered 47 new phylum level bacterial groups, naming many of them after influential microbiologists and other scientists, including ten in the Biosciences Area (with the form Candidatus Surnamebacteria). Phyla-level names have been proposed for Molecular Biophysics & Integrated Bioimaging Division’s Cheryl Kerfeld, Krishna Niyogi, and Jennifer Doudna; Environmental Genomics & Systems Biology’s Louise Glass, Kathleen Ryan, Steven Brenner, Mary Wildermuth, and Judy Wall; and the JGI’s John Vogel and Tanja Woyke. The researchers analyzed the metabolic interactions of these and other subsurface microbes to better understand their roles in ecosystem resilience. Read the full story at the Berkeley Lab News Center.
Biosciences Staff Honored with Director’s Awards
Several Biosciences Area personnel have been named as recipients of 2016 Berkeley Lab Director’s Awards. Yan Liang (Biological Systems & Engineering), Eva Nogales, and William Jagust (Molecular Biophysics & Integrated Bioimaging, MBIB) were honored with individual awards in Early Career, Scientific Achievement, and Societal Impact, respectively. Jill Fuss and Steven Yannone (MBIB) were the recipients of a team award in Technology Transfer for the launch of their company CinderBio. Jim Bristow (Biosciences Area Office, Trent Northen (Environmental Genomics & Systems Biology & Joint Genome Institute, JGI), and Susannah Tringe (JGI), along with Eoin Brodie and Peter Nico of the Earth and Environmental Sciences Area, were named in a team award in Service.
- « Previous Page
- 1
- …
- 38
- 39
- 40
- 41
- 42
- …
- 44
- Next Page »
Was this page useful?