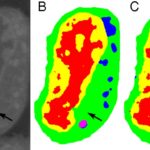
Typical machine learning methods used to analyze experimental imaging data rely on tens or hundreds of thousands of training images. But Daniël Pelt and James Sethian of Berkeley Lab’s Center for Advanced Mathematics for Energy Research Applications (CAMERA) have developed what they call a “Mixed-Scale Dense Convolution Neural Network” (MS-D) that “learns” much more quickly from a remarkably small training set. One promising application of MS-D is in understanding the internal structure and morphology of biological cells to identify, for example, differences between healthy and diseased cells. In one such project in Carolyn Larabell’s lab, the method needed data from just seven cells to determine the cell structure.
Crystallization Screen Created by Berkeley Lab Biosciences Scientists Reaches the Market
 X-ray crystallography has been the most successful technique used to solve macromolecular structures, contributing several thousand new entries to the Protein Data Bank (PDB) every year. The protein crystal is the critical starting point for X-ray data collection, and consequently, its properties are correlated with the quality of the data and the level of detail that can be extracted for a macromolecular structure. However, proteins require solutions of specific composition to form crystals for structure determination studies. These specifications are usually determined from exposing the protein to several different solutions in a crystallization screen.
X-ray crystallography has been the most successful technique used to solve macromolecular structures, contributing several thousand new entries to the Protein Data Bank (PDB) every year. The protein crystal is the critical starting point for X-ray data collection, and consequently, its properties are correlated with the quality of the data and the level of detail that can be extracted for a macromolecular structure. However, proteins require solutions of specific composition to form crystals for structure determination studies. These specifications are usually determined from exposing the protein to several different solutions in a crystallization screen.
A team of researchers in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division led by Paul Adams and Jose Henrique Pereira have developed a new crystal screen, the Berkeley Screen, with 96 conditions proven to be highly effective at producing crystals for structural determination. The Berkeley Screen is now available to the wider crystallography community commercially.
ALS Shines New Light on Evolution of Photosynthesis
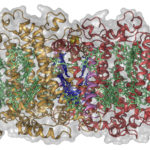
Found in muddy soils near hot springs, Heliobacterium modesticaldum is the simplest bacterium known to be able to drive photosynthesis. Its photosynthesis reaction centers are thought to resemble the earliest common ancestor of all photosynthesis complexes, which evolved around three billion years ago. Now, for the first time, a team led by researchers from Arizona State University has obtained a near-atomic resolution (2.2 Å) structure of the membrane protein at the heart of H. modesticaldum’s photosynthetic reaction center using X-ray crystallography data collected at ALS Beamline 8.2.1. The structure gives researchers a new perspective on the early evolution of photosynthesis. Read more in this ALS Science Brief.
CRISPR Target Recognition Mechanism Illuminated Using Data From ALS
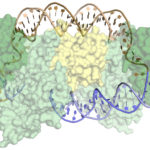
Using diffraction data obtained at Berkeley Lab’s Advanced Light Source (ALS) and at the Stanford Synchrotron Radiation Lightsource (SSRL), researchers in Biosciences’ Molecular Biophysics & Integrated Bioimaging (MBIB) division and at UC Berkeley have discovered how CRISPR-associated (Cas) proteins are able to recognize their target locations with such great specificity. X-ray crystallography was used to solve the structures of Cas1 and Cas2—responsible for DNA-snippet capture and integration—as they were bound to synthesized DNA strands designed to mimic different stages of the process. The resulting structures show how the system works in its native context as part of a bacterial immune system, and also inform the development of the CRISPR-Cas system as a general-purpose molecular recording device. Jun-Jie Liu, a joint postdoc in the labs of Jennifer Doudna and Eva Nogales, and Addison Wright, a graduate student in Doudna’s lab, were co-first authors on the paper, published in the journal Science. Read more in this ALS Science Highlight.
NIH Awards $6.5 Million for Augmenting Structural Biology Research Experience
The National Institutes of Health (NIH) has awarded $6.5 million to Berkeley Lab to integrate existing synchrotron structural biology resources to better serve researchers. The grant will establish a center based at the Lab’s Advanced Light Source (ALS) called ALS-ENABLE that will guide users through the most appropriate routes for answering their specific biological questions.
- « Previous Page
- 1
- …
- 14
- 15
- 16
- 17
- 18
- 19
- Next Page »
Was this page useful?