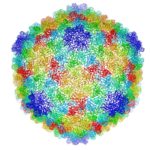
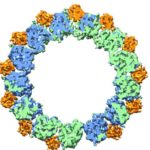
Cryo-electron microscopy is a critical tool used to advance biochemical knowledge. Now Pavel Afonine, research scientist, and Molecular Biophysics and Integrated Bioimaging Division Director Paul Adams have extended cryo-EM’s impact further by developing a new computational algorithm that was instrumental in constructing a 3-D atomic-scale model of bacteriophage P22 for the first time. Read more in the Berkeley Lab News Center.
Finding Diamonds in the Rough
New crystallography finding by JBEI and GLBRC benefits bioenergy industry
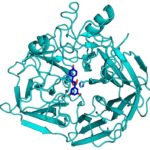 During the kraft process used to convert wood into wood pulp, the structural material lignin is partially converted into molecules like stilbene. Stilbenes are also naturally occurring in plants and some bacteria, and may play a role in plant pathogen resistance.
During the kraft process used to convert wood into wood pulp, the structural material lignin is partially converted into molecules like stilbene. Stilbenes are also naturally occurring in plants and some bacteria, and may play a role in plant pathogen resistance.
Currently, the deconstruction of plant biomass into cellulose and lignin is an expensive process. Lignin accounts for about 30 percent of plant cell wall carbon, and its conversion into chemicals or fuels could have a significant positive impact on the economics of processing lignocellulosic biomass. Enzymes capable of producing useful compounds from the breakdown of stilbenes and similar molecules could be employed for this. Collaborators from two of the Department of Energy Bioenergy Research Centers now have gained first-hand insight into how a stilbene cleaving oxygenase (SCO) carries out this unusual chemical reaction.
X-rays Capture Unprecedented Images of Photosynthesis in Action
An international team of scientists is getting closer to discovering how plants split water during photosynthesis and produce nearly all of the oxygen in our atmosphere. Thanks to unprecedented, atomic-scale images of a protein complex found in plants, algae, and cyanobacteria captured by ultrafast X-ray lasers, researchers conducted atomic-level experiments to help delineate the mechanism of this system that also yields the protons and electrons used to reduce carbon dioxide to carbohydrates later in the photosynthesis cycle. The effort to uncover the secrets of this protein complex, photosystem II, was led by Vittal Yachandra and Junko Yano in the Molecular Biophysics & Integrated Bioimaging (MBIB) Division and the team’s findings were published this week in Nature.
Workshop Focuses in on Electron Microscopy
An all-day workshop highlighting the Berkeley Lab’s capabilities in electron microscopy was held on Tuesday, October 11. Organized by Paul Adams (Biosciences Area), Peter Denes (Advanced Light Source) and Andy Minor (National Center for Electron Microscopy), the workshop highlighted recent advances in imaging a broad range of materials and biological samples at atomic, or near-atomic scales. In addition, it made evident the many opportunities that could come from integrating capabilities across the Laboratory. Paul Adams, Director of the Molecular Biophysics & Integrated Bioimaging Division, noted that the recent revolution in electron microscopy for biosciences has opened up many new avenues of research and exciting synergies with non-biosciences programs at the Lab. Read more in Glenn Roberts’ Science Short on the Berkeley Lab News Center.
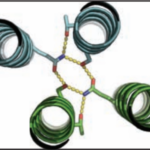
Bioscientists Validate Novel Protein Design Program
Over the course of billions of years, nature has evolved particular molecular structures that form the basis of life, such as those found in nucleic acids and proteins. Using the natural form as a springboard, University of Washington researchers have designed protein homo-oligomers, or identical interacting subunits, which can contain interchangeable hydrogen bonding modules for building different structures or functions. The team of researchers, led by David Baker at the University of Washington, included Jose Henrique Pereira, Banumathi Sankaran, and Peter Zwart of the Molecular Biophysics & Integrated Bioimaging Division (MBIB).
- « Previous Page
- 1
- …
- 6
- 7
- 8
- 9
- Next Page »
Was this page useful?