Eva Nogales, faculty scientist in Molecular Biophysics & Integrated Bioimaging (MBIB) Division and UC Berkeley professor of molecular and cell biology, led a team that captured freeze-frames of the changing shape of a huge macromolecular complex as it locks onto DNA and loads the machinery for reading the genetic code. 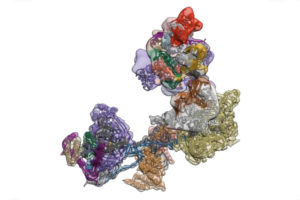 The molecule, called transcription factor IID (TFIID), is critical to transcribing genes into messenger RNA that will later be used as blueprints to make proteins. Because of its many moving parts and large size, however, TFIID’s 3D structure has been hard to capture, with the previous results being a blurred composite image of many different conformations devoid of dynamic information.
The molecule, called transcription factor IID (TFIID), is critical to transcribing genes into messenger RNA that will later be used as blueprints to make proteins. Because of its many moving parts and large size, however, TFIID’s 3D structure has been hard to capture, with the previous results being a blurred composite image of many different conformations devoid of dynamic information.
The scientists used the imaging technique cryoelectron microscopy (cryo-EM) to obtain snapshots of this structure as it engages with DNA, scans the sequence, and recruits and rearranges the appropriate proteins. High-resolution structural information like this is essential for understanding how TFIID translates the operating instructions in the genome and how it could malfunction. The new, more detailed snapshots of the molecule’s moving parts could help drug designers create therapeutics that interfere with the molecule’s structural changes in order to tweak the expression of a gene that is causing disease.
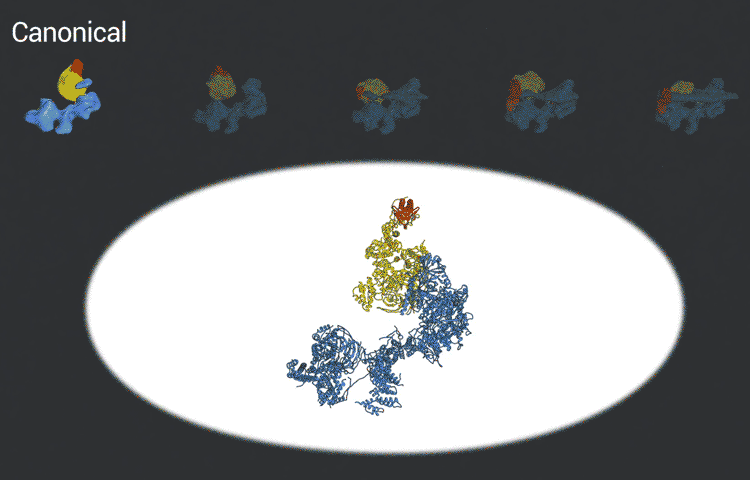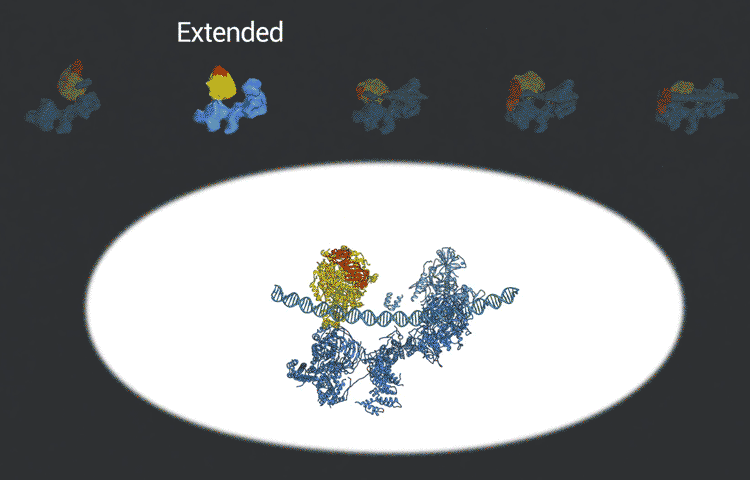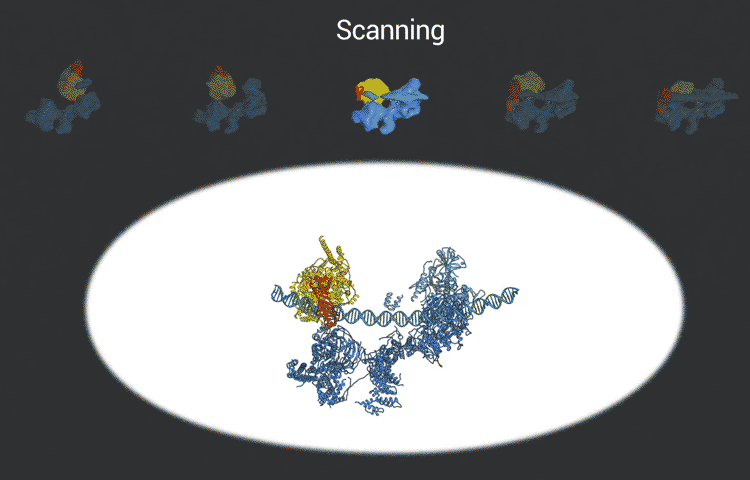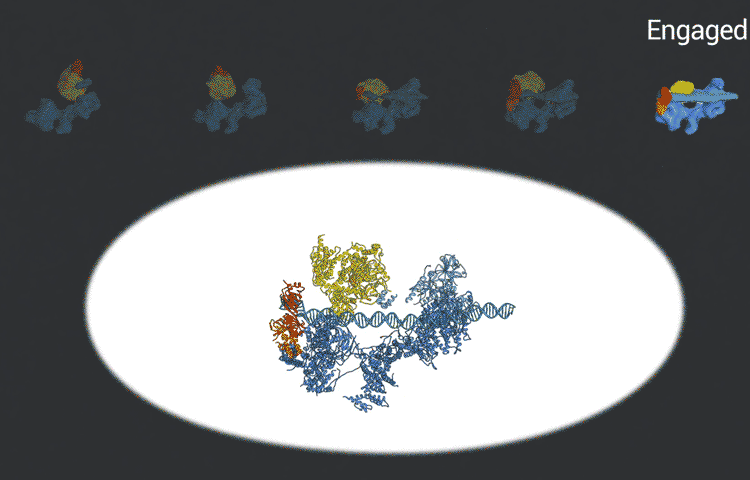
Nogales and her team, which includes co-first authors on the study, Avinash Patel and Robert Louder, both UC Berkeley graduate students and MBIB affiliates, posted their findings online in advance of print publication in the journal Science. The researchers used Phenix, a software suite for solving three-dimensional macromolecular structures developed under the direction of MBIB Division Director Paul Adams, to refine the model, as well as the resources of the National Energy Research Scientific Computing Center (NERSC), a DOE Office of Science User Facility.
Read more in the UC Berkeley news release.



