To characterize the functions of genes in newly discovered bacteria, microbiologists introduce mutations using mobile DNA snippets called transposons delivered via a vector. But finding an effective transposon mutagenesis system for a novel microorganism through trial and error can be a time-consuming and expensive endeavor.
To streamline the process, Environmental Genomics and Systems Biology (EGSB) Division researchers, led by Adam Deutschbauer and Adam Arkin, have developed a new technique to test hundreds of different transposon vector variants in parallel within what the investigators term “magic pools.”
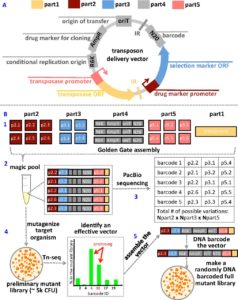
The approach was detailed in mSystems, an open access journal from the American Society for Microbiology. Hualan Liu, a postdoc in Deutschbauer’s lab, was first author on the paper.
The researchers first assembled a collection of hundreds of individual DNA vectors with different combinations of genetic material and antibiotic resistance markers. To enable the tracking of all of these different transposon vectors in parallel, a unique DNA barcode was added to each vector.
After introducing mutations to a new bacterium with the magic pool, DNA sequencing of the barcode can be used to identify the vector design that is effective for making transposon mutants. The team used this method to construct large libraries of transposon mutants in five bacteria, including three from the phylum Bacteroidetes.
Deutschbauer noted that the general strategy could also be used to accelerate the development of non-transposon-based genetic systems, such as CRISPR/Cas genome editing or plasmid-based overexpression, in microorganisms. Data describing the magic pools, the full mutant libraries, and the mutant fitness assays, as well as source code, have been made available to the research community at a dedicated portal.
Additional Berkeley Lab researchers who contributed to the work were: Morgan Price (Arkin lab); Robert Jordan Waters (Matthew Blow lab); Jayashree Ray, Hans Carlson, and Jacob Lamson (Deutschbauer lab); and Romy Chakraborty (Earth and Environmental Sciences Area).



