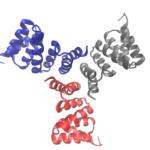
Cyclic proteins that assemble from multiple identical subunits (homo-oligomers) play key roles in many biological processes, including enzymatic catalysis and function and cell signaling. Researchers in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division worked with University of Washington’s David Baker, who led a team to design in silico and crystallize self-assembling cyclic homo-oligomer proteins.
Unexpected Key Role for Unfolded Protein Regions in DNA Break Repair Defined
Structurally Integrated Biology for Life Sciences (SIBYLS) beamline researchers, led by research scientist Michal Hammel of the Molecular Biophysics & Integrated Bioimaging (MBIB) Division, used X-ray scattering to define an unexpected key role for unfolded protein regions in DNA break repair to allow regulation plus access to DNA ends. The concept of DNA break repair as a flexibly-linked dynamic complex, as opposed to a linear pathway, suggests new approaches to targeting DNA repair for selectively killing cancer cells due to their high levels of DNA instability. Their findings were published in a recent cover article of the Journal of Biological Chemistry.
The SIBYLS beamline of the Advanced Light Source at Berkeley Lab, directed by MBIB’s senior scientist John Tainer, is optimized for both small-angle X-ray scattering (SAXS) and macromolecular crystallography (MX), making it unique among the world’s mostly SAXS or MX dedicated beamlines.
Exploring the Repeat-Protein Universe
Naturally occurring proteins—chains of amino acids that fold into functional, three-dimensional shapes—are believed to represent just a small fraction of the universe of all possible permutations of amino-acid sequences and folds.  How can we begin to systematically sift through those permutations to find and engineer from scratch (de novo) proteins with the characteristics desired for medical, environmental, and industrial purposes? To address this question, a team led by researchers from the Institute for Protein Design at the University of Washington have published a landmark study that used both protein crystallography (Beamlines 8.2.1 in the Berkeley Center for Structural Biology and 8.3.1) and small-angle x-ray scattering (SAXS; SIBYLS Beamline) at the ALS to validate the computationally designed structures of novel proteins with repeated motifs. The results show that the protein-folding universe is far larger than realized, opening up a wide array of new possibilities for biomolecular engineering. Read the ALS Science Highlight.
How can we begin to systematically sift through those permutations to find and engineer from scratch (de novo) proteins with the characteristics desired for medical, environmental, and industrial purposes? To address this question, a team led by researchers from the Institute for Protein Design at the University of Washington have published a landmark study that used both protein crystallography (Beamlines 8.2.1 in the Berkeley Center for Structural Biology and 8.3.1) and small-angle x-ray scattering (SAXS; SIBYLS Beamline) at the ALS to validate the computationally designed structures of novel proteins with repeated motifs. The results show that the protein-folding universe is far larger than realized, opening up a wide array of new possibilities for biomolecular engineering. Read the ALS Science Highlight.
Was this page useful?