On a social network like Facebook, each user (person or organization) is represented as a node and the connections (relationships and interactions) between them are called edges. By analyzing these connections, researchers can learn a lot about each user. In biology, similar graph-clustering algorithms can be used to understand the proteins that perform most of life’s functions. Today, advanced high-throughput technologies allow researchers to capture hundreds of millions of proteins, genes and other cellular components at once and in a range of environmental conditions. Clustering algorithms are then applied to these datasets to identify patterns and relationships that may point to structural and functional similarities. Though these techniques have been widely used for more than a decade, they cannot keep up with the torrent of biological data being generated by next-generation sequencers and microarrays. In fact, very few existing algorithms can cluster a biological network containing millions of nodes (proteins) and edges (connections). That’s why a team of Berkeley Lab researchers, including scientists at the Joint Genome Institute (JGI), took one of the most popular clustering approaches in modern biology—the Markov Clustering (MCL) algorithm—and modified it to run quickly, efficiently and at scale on distributed-memory supercomputers. In a test case, their high-performance algorithm—called HipMCL—achieved a previously impossible feat: clustering a large biological network containing about 70 million nodes and 68 billion edges in a couple of hours, using approximately 140,000 processor cores on the National Energy Research Scientific Computing Center’s (NERSC) Cori supercomputer. A paper describing this work was recently published in the journal Nucleic Acids Research. Read the whole story on the Berkeley Lab Computational Research Division site.
Impact of Environmental Changes on Microbes in Arctic Soils
As the Arctic continues to warm at about twice the rate of the rest of the world, scientists expect its frozen soils—known as permafrost—to thaw, activating microbes capable of decomposing soil and releasing carbons and other nutrients to the atmosphere and water. Berkeley Lab scientists in the Earth and Environmental Systems Area (EESA), led by microbial ecologist Neslihan Taş, set out to learn more about how Arctic soil microbes can contribute to greenhouse gas emissions under a warming climate. Taş’s research team collaborated with Susannah Tringe, Deputy for User Programs at the Joint Genome Institute (JGI), to conduct their study, funded by the Department of Energy’s Office of Biological and Environmental Research (BER). The results were published in Nature Communications.
Area Scientists to Present Talks at 2018 AAAS Annual Meeting
Several Berkeley Lab scientists will present talks at the 72nd annual meeting of the American Association for the Advancement of Science (AAAS), the world’s largest general scientific society, to be held February 15 through 19 in Austin, Texas. Among them are four representing the Biosciences Area: Mary Maxon, Blake Simmons, Trent Northen, and Susannah Tringe.
JGI Helps Track Microbial Diversity Through the Terrestrial Subsurface
In collaboration with a team led by longtime collaborator Jill Banfield of the University of California, Berkeley and Cathy Ryan of the University of Calgary in Canada, JGI researchers investigated samples collected at Utah’s Crystal Geyser over one of its complex, five-day eruption cycles. Genome-resolved metagenomics, single-cell genomics, and geochemical analyses were integrated to show that samples taken during each phase contain microbial communities that are distinctive in terms of both composition and metabolic function. The report was published January 29, 2018 in Nature Microbiology. Read the full story on the JGI website.
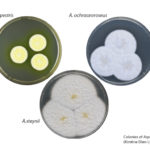
JGI, JBEI Present First Results of Genus-Wide Aspergillus Project
Found in microbial communities around the world, Aspergillus fungi are pathogens, decomposers, and important sources of biotechnologically-important enzymes. In a study published ahead the week of January 8, 2018 in the Proceedings of the National Academy of Sciences, a team led by researchers at the Technical University of Denmark (DTU), the Joint Genome Institute (JGI), and the DOE’s Joint BioEnergy Institute (JBEI) report the first results of a long-term plan to sequence, annotate and analyze the genomes of 300 Aspergillus fungi. These findings are a proof of concept of novel methods to functionally annotate genomes in order to more quickly identify genes of interest. Read more on the JGI website.
- « Previous Page
- 1
- …
- 31
- 32
- 33
- 34
- 35
- …
- 46
- Next Page »
Was this page useful?