In Nature, researchers in Berkeley Lab’s Biosciences Area, the Gladstone Institutes, and the Chan-Zuckerberg Biohub presented nearly 61,000 microbial genomes that were computationally reconstructed from 3,810 publicly available human gut metagenomes, which are datasets of all the genetic material present in a microbiome sample. The metagenome-assembled genomes (MAGs) included 2,058 previously unknown species, bringing the number of known human gut species to 4,558 and increasing the phylogenetic diversity of sequenced gut bacteria by 50 percent. This work helps answer the question of why certain microbes have not been cultivated in the lab. Read more in the Berkeley Lab News Center.
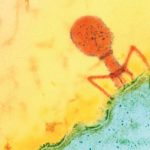
Megaphages Discovered in Human Gut Bacteria
While sequencing gut bacteria from people in Bangladesh, Berkeley Lab’s Jillian Banfield discovered phages, viruses that infect and reproduce inside bacteria, twice as big as any previously found in humans. She and her colleagues found the snippets of megaphage DNA in a CRISPR segment of one type of bacteria, Prevotella, that is uncommon in people eating a high-fat, low-fiber Westernized diet. Banfield and her team named the clade of megaphages “Lak phage” after the Laksam Upazila area of Bangladesh where they were found.
Updating the Genome Data Sharing Agreement
Nearly 20 years ago in Fort Lauderdale, Fla., genome data producers and data users came to an accord on the use of genome sequencing data released to the public domain. In particular, they agreed that the data was freely available for use and access by the scientific community before those data are used for publication. The Fort Lauderdale Agreement did not include defined policies on data usage, and has led to years of debate, such as whether or not there was a tacit acknowledgement that data generators would have the right of first publication on the data they produced and freely shared.
In a policy paper published January 25, 2019 in Science, 50 coauthors, with 54 unique affiliations from 18 countries, call for a “clear policy that protects public data from restrictions.” The international consortium includes Nikos Kyrpides of the Biosciences Area’s Environmental Genomics & Systems Biology (EGSB) Division at Lawrence Berkeley National Laboratory (Berkeley Lab).
JGI Researchers Lead Community Effort for Defining Virus Data Quality
“Viruses are critical components of every microbial ecosystem. The JGI is especially interested in developing standards for virus genomes because we generate much of this data ourselves,” said JGI research scientist and first author Simon Roux.
As more and more researchers continue to assemble new genome sequences of uncultivated viruses, JGI researchers led a community effort to develop guidelines and best practices for defining virus data quality.
JGI Helps Develop Model System for Perennial Grasses
Reported in Nature Communications, a team led by Tom Juenger at the University of Texas (UT) at Austin and including JGI researchers describe the culmination of nearly a decade of work to develop genomic resources for drought tolerance in perennial grasses. The team aims to apply the resources developed for Panicum hallii towards stress tolerance improvement in its more complex relative, the candidate bioenergy crop, switchgrass.
Through the JGI’s Community Science Program, JGI sequenced and assembled near-complete genomes of P. hallii var. hallii (99.2% complete) and P. hallii var. filipes (94.8% complete) and resequenced a host of natural collections from across the species range. With these high-quality reference genomes for P. hallii, researchers can identify and characterize the regulatory elements that influence adaptation and tolerance to stressors such as drought. This information can be applied toward improving crop yields in other grasses. Click here to read the JGI science highlight.
- « Previous Page
- 1
- …
- 24
- 25
- 26
- 27
- 28
- …
- 46
- Next Page »
Was this page useful?