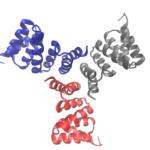
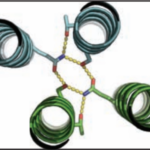
Cyclic proteins that assemble from multiple identical subunits (homo-oligomers) play key roles in many biological processes, including enzymatic catalysis and function and cell signaling. Researchers in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division worked with University of Washington’s David Baker, who led a team to design in silico and crystallize self-assembling cyclic homo-oligomer proteins.
Designing Protein Cavities from Curved Beta Sheets
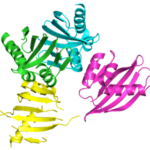
Curved beta sheets are important for the architecture of protein cavities, such as enzyme active sites and ligand-binding pockets. Beginning by analyzing classic protein formations and running folding simulations, University of Washington (UW) researchers under the leadership of David Baker designed six protein folds inspired by naturally occurring protein superfamilies. A research report published in the January 13 issue of Science describes how a multi-institutional team of scientists compared the predicted models to physical structures of these designed proteins.
Three Is Not a Crowd: Designed Metalloprotein Trimer Provides Stable Platform for Further Development
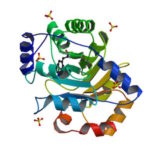 University of Washington (UW) researchers have designed a novel protein with properties that could lead to the generation of new photoactive proteins. This three-fold symmetric, self-assembling protein homotrimer contains a highly stable noncanonical amino acid. Noncanonical amino acids are not found among the 20 encoded amino acids in the body and can contain modifications to allow for new functionality. In this case, this amino acid contains a bipyridine group that chelates metal, thereby introducing new photochemical properties into the protein interface, and nucleating the formation of the homotrimer.
University of Washington (UW) researchers have designed a novel protein with properties that could lead to the generation of new photoactive proteins. This three-fold symmetric, self-assembling protein homotrimer contains a highly stable noncanonical amino acid. Noncanonical amino acids are not found among the 20 encoded amino acids in the body and can contain modifications to allow for new functionality. In this case, this amino acid contains a bipyridine group that chelates metal, thereby introducing new photochemical properties into the protein interface, and nucleating the formation of the homotrimer.
An article published last month in PNAS describes this work from a team of scientists led by David Baker at UW, which included Jose Henrique Pereira, Banumathi Sankaran, and Peter Zwart of the Molecular Biophysics & Integrated Bioimaging Division (MBIB). The MBIB scientists developed the crystal screen that was used to crystallize the novel protein and performed X-ray crystallography on Beamline 8.2.1 in the Berkeley Center for Structural Biology at the Advanced Light Source. Their X-ray crystallographic analysis of the homotrimer showed that the design process had near-atomic-level accuracy, demonstrating that computational protein design together with the utilization of noncanonical amino acids could be used to generate novel protein functions. These methods could be used to develop new therapeutics, biomaterials, and metalloproteins with useful optical or photochemical properties.
Finding Diamonds in the Rough
New crystallography finding by JBEI and GLBRC benefits bioenergy industry
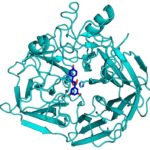 During the kraft process used to convert wood into wood pulp, the structural material lignin is partially converted into molecules like stilbene. Stilbenes are also naturally occurring in plants and some bacteria, and may play a role in plant pathogen resistance.
During the kraft process used to convert wood into wood pulp, the structural material lignin is partially converted into molecules like stilbene. Stilbenes are also naturally occurring in plants and some bacteria, and may play a role in plant pathogen resistance.
Currently, the deconstruction of plant biomass into cellulose and lignin is an expensive process. Lignin accounts for about 30 percent of plant cell wall carbon, and its conversion into chemicals or fuels could have a significant positive impact on the economics of processing lignocellulosic biomass. Enzymes capable of producing useful compounds from the breakdown of stilbenes and similar molecules could be employed for this. Collaborators from two of the Department of Energy Bioenergy Research Centers now have gained first-hand insight into how a stilbene cleaving oxygenase (SCO) carries out this unusual chemical reaction.
Bioscientists Validate Novel Protein Design Program
Over the course of billions of years, nature has evolved particular molecular structures that form the basis of life, such as those found in nucleic acids and proteins. Using the natural form as a springboard, University of Washington researchers have designed protein homo-oligomers, or identical interacting subunits, which can contain interchangeable hydrogen bonding modules for building different structures or functions. The team of researchers, led by David Baker at the University of Washington, included Jose Henrique Pereira, Banumathi Sankaran, and Peter Zwart of the Molecular Biophysics & Integrated Bioimaging Division (MBIB).
Was this page useful?