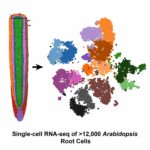
Researchers led by Diane Dickel have successfully adapted an open-source RNA analysis platform to study gene expression in individual plant cells. The method, called Drop-seq, was developed at Harvard Medical School in 2015 and had previously been used only in animal cells. Dickel and her colleagues at the DOE Joint Genome Institute (JGI) teamed up with researchers from UC Davis who had perfected a protoplasting technique for root tissue from Arabidopsis thaliana (mouse-ear cress). After preparing samples of more than 12,000 Arabidopsis root cells, the group was thrilled when the Drop-seq process went smoother than expected. Their results were published in Cell Reports.
A Synthetic Nanofactory Inspired by Nature
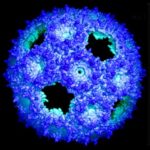
Researchers at Berkeley Lab and Michigan State University (MSU), led by Cheryl Kerfeld, have created a genetically engineered bacterial microcompartment (BMC) shell based on natural structures and the principles of protein evolution. The new shell is smaller and simpler, made of only a single designed protein (natural BMCs are made of up to three), making it easier to work with in the lab.
Pam Ronald Elected to National Academy of Sciences
Pam Ronald, faculty scientist in EGSB and scientific lead of plant pathology at the Joint BioEnergy Institute (JBEI), was elected to the National Academy of Sciences in recognition of her distinguished and continuing achievements in original research. She joins 100 scientists and engineers from the U.S. and 25 from across the world as new lifelong members and foreign associates.
Regulation of Algal Photosynthesis and Metabolism
The unicellular green alga Chromochloris zofingiensis has the ability to shift metabolic modes from photoautotrophic (synthesizing food using light as energy source) to heterotrophic (obtaining food and energy from exogenous sources) in response to carbon source availability in the light. It also has the capacity—under certain conditions—to produce high amounts of commercially relevant bioproducts: notably, the ketocarotenoid astaxanthin, used in feed, cosmetics, and as a nutraceutical, and triacylglycerol (TAG) biofuel precursors.
Understanding how photosynthesis and metabolism are regulated in algae could, via bioengineering, enable scientists to reroute metabolism toward beneficial bioproducts for energy, food, and human health. To that end, Berkeley Lab Biosciences researchers used C. zofingiensis as a simple algal model system to investigate conserved eukaryotic sugar responses, as well as mechanisms of thylakoid breakdown and biogenesis in chloroplasts.
First Look at New Light Absorbing Protein
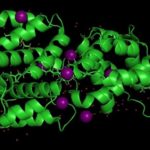
The Helical Carotenoid Protein 2 (HCP2) protein is an ancestor of proteins that are known to protect against damage caused by excess light exposure. Researchers in the laboratory of Cheryl Kerfeld, guest faculty in the Environmental Genomics & Systems Biology (EGSB) Division, are the first to structurally and biophysically analyze a protein from the HCP family. This HCP protein family was discovered recently by Kerfeld and the members of her lab, who are based in EGSB and at Michigan State University (MSU). To solve the molecular structure of HCP2, X-ray diffraction was measured at beam line 5.0.2 in the Berkeley Center for Structural Biology of the Advanced Light Source (ALS). The structure was refined using Phenix, a software suite for automated determination of molecular structures developed under the direction of Paul Adams, Molecular Biophysics and Integrated Bioimaging Division Director. Read more in the MSU-DOE Plant Research Laboratory news story.
- « Previous Page
- 1
- …
- 22
- 23
- 24
- 25
- 26
- …
- 46
- Next Page »
Was this page useful?