JBEI researchers develop efficient and affordable method for plant DNA assembly
Researchers at the U.S. Department of Energy (DOE)’s Joint BioEnergy Institute (JBEI) in collaboration with Berkeley Lab’s Environmental Genomics & Systems Biology Division and the DOE Joint Genome Institute developed a versatile system (named jStack) which utilizes yeast homologous recombination to efficiently assemble DNA into plant transformation vectors. The new approach will impact plant engineering for the bioenergy, agricultural and pharmaceutical industries.
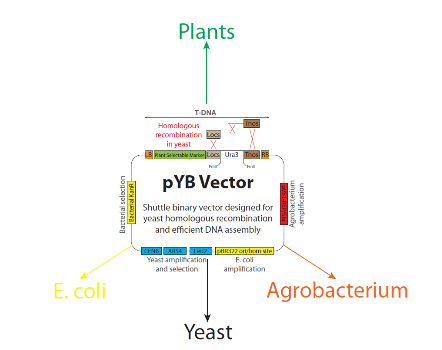 Although synthetic biology has provided solutions to many societal challenges, little research has been devoted to advancing synthetic biology in plants. Microbes, such as yeast and Escherichia coli (E. coli), have received much of the attention in developing synthetic biology tools due to their fast generation time and the ease of working with these organisms in laboratories. A shortage of characterized DNA parts, along with the difficulty of efficiently assembling multiple and large fragments of DNA into plant transformation vectors, has limited progress in studying and engineering plants to the same degree as their microbial counterparts.
Although synthetic biology has provided solutions to many societal challenges, little research has been devoted to advancing synthetic biology in plants. Microbes, such as yeast and Escherichia coli (E. coli), have received much of the attention in developing synthetic biology tools due to their fast generation time and the ease of working with these organisms in laboratories. A shortage of characterized DNA parts, along with the difficulty of efficiently assembling multiple and large fragments of DNA into plant transformation vectors, has limited progress in studying and engineering plants to the same degree as their microbial counterparts.
A new paper entitled “A robust gene-stacking method utilizing yeast assembly for plant synthetic biology” published in Nature Communications (published online on October 26), describes a new platform, jStack, which provides a powerful synthetic biology tool for a wide range of plant researchers to improve gene- and trait-stacking efforts to support fundamental research and engineering agriculturally relevant crops.
“We can only advance agriculture, bioenergy and crop sustainability if we lay down a solid foundation in plant synthetic biology,” said Dominique Loqué, Principal Investigator for this study and Director of Cell Wall Engineering at the Joint Bioenergy Institute. “We believe that the jStack platform is a step in the right direction as it provides a robust and comprehensive tool for plant DNA assembly.”
Patrick Shih, the first author of this paper and a postdoctoral researcher in Loqué’s lab developed a versatile approach which utilizes yeast homologous recombination to efficiently assemble DNA into plant transformation vectors. Previously, yeast homologous recombination approaches have been utilized to achieve once-thought-to-be-impossible synthetic biology feats such as the de novo assembly of whole bacterial genomes. In this study, JBEI researchers have refashioned this approach to drastically increase the efficiency and flexibility of assembling large DNA cassettes into plant expression vectors.
Another important result from this research was building a library of plant-specific DNA parts (i.e., promoters, genes, terminators). Most of the time, gene assembly requires the use of characterized building blocks, however there is a shortage of those characterized DNA parts that are plant-specific and compatible for Type IIS restriction endonuclease-mediated assembly. JBEI researchers have tackled this shortage by generating a library of 115 plant-specific DNA parts that follow a common syntax adopted by several international plant laboratories and which have been made publicly available to other plant researchers through JBEI’s Inventory of Composable Elements (ICE), a cloud-based freely open-source DNA/strains/seeds repository).
“With this study we have been able to demonstrate how our tools and resources can be applied to introduce complex heterologous pathways into plants,” said Patrick Shih. “Moreover, we showcase the potential of our approach in assembling chromosomal regions and building synthetic gene clusters to enable the stacking and shuffling of diseases resistance traits between crop plants. Ultimately it will have great impact on fundamental research and engineering agriculturally relevant crops” added Loqué.



