Through a decade-long time-series study recently reported in Microbiome, a team led by Rick Cavicchioli of Australia’s University of New South Wales and including JGI researchers found that light-harvesting bacteria in Antarctica’s Ace Lake defy the norm when it comes to nutrient cycling in polar regions. While microbial populations in other Antarctic lakes and the nearby Southern Ocean shift from phototrophs to archaea when sunlight becomes scant, Ace Lake’s bacteria instead go through a boom and bust cycle aligned with light availability. Read more on the JGI website.
JGI’s Simon Roux Receives DOE Early Career Research Award
Congratulations to JGI research scientist Simon Roux, who is among the Berkeley Lab scientists awarded significant funding for research through the DOE Office of Science Early Career Research Program (ECRP). A total of 76 ERCP recipients were selected this year. The scientists are each expected to receive grants of up to $2.5 million over five years to cover salary and research expenses.
JGI Overhauls Perception of Inovirus Diversity
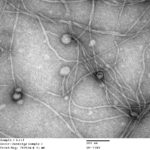
Inoviruses are filamentous viruses with small, single-stranded DNA genomes and a unique chronic infection cycle. In Nature Microbiology, a team led by DOE Joint Genome Institute (JGI) researchers applied machine learning to publicly available microbial genomes and metagenomes to search for inoviruses. The search tool combed through more than 70,000 microbial and metagenome datasets, ultimately identifying more than 10,000 inovirus-like sequences compared to the 56 previously known inovirus genomes. The results revealed inoviruses are in every major microbial habitat—including soil, water, and humans—around the world.
“We’re not sure why we systematically manage to miss them; maybe it’s due to the way we currently isolate and extract viruses,” said the study’s lead author Simon Roux, a JGI research scientist in the Environmental Genomics group. Click here to read the full story on the JGI site.
JGI Researchers Featured in mSystems Special Issue
Three Joint Genome Institute researchers are among the authors who offered perspectives on what the next five years of innovation could look like for a special issue of the journal mSystems. In one article, Micro-Scale Applications head Rex Malmstrom and Metagenome Program head Emiley Eloe-Fadrosh outline more targeted approaches to reconstruct individual microbes in an environmental sample. In a separate article, research scientist Simon Roux, a member of Eloe-Fadrosh’s Environmental Genomics group, makes a pitch for readers to get involved in the developing field of virus ecogenomics.
Read more from JGI.
Biosciences Area FY19 LDRD Projects
The projects of 13 Biosciences Area scientists and engineers received funding through the FY19 Laboratory Directed Research and Development (LDRD) program. The funded projects span a diverse array of topics and approaches including the harnessing of microbiome data to uncover patterns of mutualism, evaluating radiobiological effects of laser-accelerated ion beams, improving bioenergy yield under drought stress, and the application of machine learning in tomogram segmentation. Lab-wide, 89 projects were selected from a field of 158 proposals. Biosciences Area efforts account for 15.07 percent of the $22.2 million allocated.
Was this page useful?