Six proposals have been selected to participate in a new partnership between two DOE user facilities at Lawrence Berkeley National Laboratory through the FICUS initiative. The expertise and capabilities available at JGI and NERSC will help researchers explore the wealth of genomic and metagenomic data generated worldwide through access to supercomputing resources and computational science experts to accelerate discoveries. Read more on the DOE JGI website.
Six proposals have been selected to participate in a new partnership between two DOE user facilities at Lawrence Berkeley National Laboratory through the FICUS initiative. The expertise and capabilities available at JGI and NERSC will help researchers explore the wealth of genomic and metagenomic data generated worldwide through access to supercomputing resources and computational science experts to accelerate discoveries. Read more on the DOE JGI website.
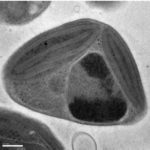
DOE JGI Helps Find How Red Alga Thrives in Intertidal Zone
As part of a 50-member team led by University of Maine, Carnegie Institution for Science, and East Carolina University researchers, the DOE Joint Genome Institute (DOE JGI) sequenced, assembled and annotated the genome of the red alga Porphyra umbilicalis. The algal genome offers insights into the organism’s stress-tolerance mechanisms and how that impacts its ability to fix carbon. Read more on the JGI website.
DOE JGI Helps Develop New Technology to Access Microbial Dark Matter
Miniaturized, microfluidic-based metagenomics technology developed under the aegis of U.S. Department of Energy Joint Genome Institute’s (DOE JGI’s) Emerging Technology Opportunity Program (ETOP) has enabled researchers to shine a light on so-called microbial “dark matter”—the majority of the planet’s microbial diversity that remains uncultivated. Researchers from Stanford University demonstrated the efficacy of the new technique, extracting 29 novel microbial genomes from Yellowstone hot spring samples while still preserving single-cell resolution to enable accurate analysis of genome function and abundance. Read more in this JGI Science Highlight.
Tiny Green Algae Reveal Large Genomic Variation
A decade after the complete representative genomes of three Ostreococcus groups were sequenced, an international team of researchers including scientists at the Department of Energy Joint Genome Institute (DOE JGI) have resequenced and analyzed the genomes of 13 members of a natural population from the northwest Mediterranean Sea. The analysis revealed that the O. tauri population is larger than anticipated, with high genetic and phenotypic diversity that is influenced by the algae’s natural resistance to ocean viruses. Understanding the genetic variability of various Ostreococcus strains will help researchers understand how environmental changes affect their abundance and ability to photosynthesize. The work was reported in Science Advances. Read more in this DOE JGI Science Highlight.
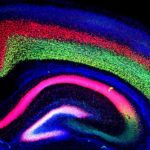
Researchers Collaborate to Create, Characterize Mouse Model of Autism
Using a genetic mouse model developed at Berkeley Lab with CRISPR/Cas9 editing technology, University of California, Davis, researchers led by Alexander Nord have examined the developmental impact of a specific mutation found in some rare cases of autism spectrum disorder (ASD). The project began while Nord—now an assistant professor with the Center for Neuroscience at UC Davis—was a postdoc in the Mammalian Functional Genomics Laboratory of Berkeley Lab co-authors Axel Visel, Len Pennacchio, and Diane Dickel. In a paper published June 26 in the journal Nature Neuroscience, the researchers report that mice with a loss-of-function mutation in one copy of the CHD8 gene have increased brain volume and cognitive impairment, similar to that seen in people with the same mutation. CHD8 encodes a protein responsible for packaging DNA in cells, which in turn controls gene expression during development. Read more from UC Davis.
- « Previous Page
- 1
- …
- 35
- 36
- 37
- 38
- 39
- …
- 46
- Next Page »
Was this page useful?