Through the JGI’s Emerging Technologies Opportunity Program (ETOP), researchers have developed and improved upon a dereplication, aggregation and scoring tool (DAS Tool) that combines existing DNA sequence binning algorithms, allowing them to reconstruct more near-complete genomes from soil metagenomes compared to other methods.
JGI Helps Identify Novel Archaeal Lineage found in Yellowstone
For decades, longtime JGI collaborator Bill Inskeep of Montana State University has been conducting microbial field studies at Yellowstone National Park. In Nature Microbiology, he and his team describe a candidate phylum-level lineage of aerobic archaea found in iron-oxide microbial mats dubbed Marsarchaeota. Through the JGI’s Community Science Program, the team used metagenome assemblies, transcriptomes and single amplified genomes from samples collected from several locations to thoroughly characterize the archaeal lineage, information that they believe will lend insights into discussions on the origin of archaea. The discovery of aerobic, thermophilic Marsarchaeota in these microbial mats provides the team with clues on how early life evolved on Earth as iron is believed to have played a key role in redox processes important in the formation and evolution of early life on Earth. Read the whole story on the JGI website.
New Workflow Accelerates Experiment-based Gene Function Assignment
While advances in sequencing technologies have enabled researchers to access the genomes of thousands of microbes and make them publicly available, the task of assigning functions to the genes uncovered has lagged behind due to the limited capacity of functional analysis approaches. To help overcome this bottleneck, Berkeley Lab researchers, led by Adam Arkin and Adam Deutschbauer in Biosciences’ Environmental Genomics and Systems Biology (EGSB) Division and Matthew Blow at the DOE Joint Genome Institute (JGI), have developed a workflow that enables large-scale, genome-wide assays of gene importance across many conditions.
Biosciences Area FY18 LDRD Projects
The projects of 13 Biosciences Area scientists and engineers received funding through the FY18 Laboratory Directed Research and Development (LDRD) program. These projects span a diverse array of topics and approaches including the study of microbiomes in relation to patterns of mutualism, crop productivity, and gut health; synthetic biology for engineering biosurfactant production and energy conversion pathways; and the application of technologies such as machine learning, high-resolution optical microscopy, and single-cell transcriptomics. Together, these efforts account for 18.75 percent of the $20 million allocated. Lab-wide, 74 projects were selected from a field of 215.
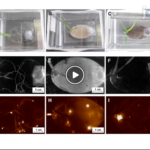
Northen Lab Publishes Video Protocol for Building EcoFABs
Scientists in Trent Northen’s groups in Environmental Genomics and Systems Biology (EGSB) and Metabolomics Technology at the DOE Joint Genome Institute (JGI) have published detailed video protocols for creating fabricated ecosystems, or EcoFABs, in the Journal of Visualized Experiments (JOVE). These laboratory-scale controlled habitats, constructed using widely available 3D printing technologies, enable mechanistic studies of plant-microbe interactions within specific environmental conditions. The published protocols serve as a starting point for other researchers, ideally helping to create standardized experimental systems for investigating plant-microbe interactions. The video component of this article can be found here.
- « Previous Page
- 1
- …
- 29
- 30
- 31
- 32
- 33
- …
- 46
- Next Page »
Was this page useful?