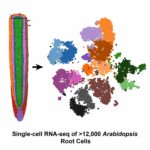
Researchers led by Diane Dickel have successfully adapted an open-source RNA analysis platform to study gene expression in individual plant cells. The method, called Drop-seq, was developed at Harvard Medical School in 2015 and had previously been used only in animal cells. Dickel and her colleagues at the DOE Joint Genome Institute (JGI) teamed up with researchers from UC Davis who had perfected a protoplasting technique for root tissue from Arabidopsis thaliana (mouse-ear cress). After preparing samples of more than 12,000 Arabidopsis root cells, the group was thrilled when the Drop-seq process went smoother than expected. Their results were published in Cell Reports.
JGI Researchers Featured in mSystems Special Issue
Three Joint Genome Institute researchers are among the authors who offered perspectives on what the next five years of innovation could look like for a special issue of the journal mSystems. In one article, Micro-Scale Applications head Rex Malmstrom and Metagenome Program head Emiley Eloe-Fadrosh outline more targeted approaches to reconstruct individual microbes in an environmental sample. In a separate article, research scientist Simon Roux, a member of Eloe-Fadrosh’s Environmental Genomics group, makes a pitch for readers to get involved in the developing field of virus ecogenomics.
Read more from JGI.
Regulation of Algal Photosynthesis and Metabolism
The unicellular green alga Chromochloris zofingiensis has the ability to shift metabolic modes from photoautotrophic (synthesizing food using light as energy source) to heterotrophic (obtaining food and energy from exogenous sources) in response to carbon source availability in the light. It also has the capacity—under certain conditions—to produce high amounts of commercially relevant bioproducts: notably, the ketocarotenoid astaxanthin, used in feed, cosmetics, and as a nutraceutical, and triacylglycerol (TAG) biofuel precursors.
Understanding how photosynthesis and metabolism are regulated in algae could, via bioengineering, enable scientists to reroute metabolism toward beneficial bioproducts for energy, food, and human health. To that end, Berkeley Lab Biosciences researchers used C. zofingiensis as a simple algal model system to investigate conserved eukaryotic sugar responses, as well as mechanisms of thylakoid breakdown and biogenesis in chloroplasts.
Biosciences Area FY19 LDRD Projects
The projects of 13 Biosciences Area scientists and engineers received funding through the FY19 Laboratory Directed Research and Development (LDRD) program. The funded projects span a diverse array of topics and approaches including the harnessing of microbiome data to uncover patterns of mutualism, evaluating radiobiological effects of laser-accelerated ion beams, improving bioenergy yield under drought stress, and the application of machine learning in tomogram segmentation. Lab-wide, 89 projects were selected from a field of 158 proposals. Biosciences Area efforts account for 15.07 percent of the $22.2 million allocated.
Seeing the Web of Microbes
Understanding nutrient flows within microbial communities is important to a wide range of fields, including medicine, bioremediation, carbon sequestration, and sustainable biofuel development. Now, researchers have built the Web of Microbes, a unique database and visualization tool that lets scientists quickly see patterns in microbial food webs. The tool, which was created through a collaboration among biologists and computational researchers at Berkeley Lab, NERSC, and the Joint Genome Institute (JGI), offers insights that other metabolic pathway databases cannot. “While most existing databases focus on metabolic pathways or identifications, the Web of Microbes is unique in displaying information on which metabolites are consumed or released by an organism in an environment such as soil,” said Suzanne Kosina, lead author on the paper and senior research associate in Biosciences’ Environmental Genomics and Systems Biology (EGSB) Division.
Read more on the Computing Sciences website.
- « Previous Page
- 1
- …
- 23
- 24
- 25
- 26
- 27
- …
- 46
- Next Page »
Was this page useful?