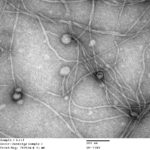
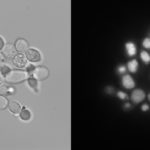
An international team of microbiologists and genomicists, led by the DOE Joint Genome Institute (JGI), has invented a genetic engineering tool to simplify the study of secondary metabolites produced by microbes. These compounds mediate internal and external messaging, self-defense, and chemical warfare, and are the basis for hundreds of valuable agricultural, industrial, and medical products. The tool, called CRAGE, will help fill significant gaps in our understanding of how microbes interact with their surroundings and evolve.
Congratulations to Biosciences Area Director’s Award Recipients
Several Biosciences Area personnel are among the 2019 Berkeley Lab Director’s Awards honorees. This annual program recognizes outstanding contributions by employees to all facets of Lab activities. A complete list of winners can be found here. The Director’s Achievement Awards ceremony will take place on November 15 at 4 PM in the Building 50 Auditorium. All staff are invited and the event will be streamed live.
JGI Helps Illuminate How a Fungus Fuels Tree Growth
The fungus Mortierella elongata enjoys a close association with the fast-growing poplar tree (Populus trichocarpa), a potential biofuel feedstock. Scientists routinely spot the fungus near the tree or in between its root cells. To better understand their relationship, a team of scientists has studied the effect of the fungus on the tree’s physical traits and gene expression, finding the fungus induces some notable metabolic changes. Click here to read the science highlight on the JGI website.
JGI Overhauls Perception of Inovirus Diversity
Inoviruses are filamentous viruses with small, single-stranded DNA genomes and a unique chronic infection cycle. In Nature Microbiology, a team led by DOE Joint Genome Institute (JGI) researchers applied machine learning to publicly available microbial genomes and metagenomes to search for inoviruses. The search tool combed through more than 70,000 microbial and metagenome datasets, ultimately identifying more than 10,000 inovirus-like sequences compared to the 56 previously known inovirus genomes. The results revealed inoviruses are in every major microbial habitat—including soil, water, and humans—around the world.
“We’re not sure why we systematically manage to miss them; maybe it’s due to the way we currently isolate and extract viruses,” said the study’s lead author Simon Roux, a JGI research scientist in the Environmental Genomics group. Click here to read the full story on the JGI site.
JGI Helps Optimize Microbial Oil Production
Yarrowia lipolytica is a tantalizing microbial factory, capable of turning abundant biomass carbohydrates—glucose and xylose—into fatty acids that could be used for renewable fuel. In fact, lipids can make up more than 90 percent of the microbe’s dry weight.
In order to better harness these gobs of energy, a team of researchers sought to disrupt every gene in the genome with CRISPR-Cas9 in order to interrogate each one’s function. However, the team first needed to fix a defect in the approach: CRISPR-Cas9 relies on RNA with a short guiding sequence (20 bp) that directs the endonuclease where to cut, but these “single guide” RNAs (sgRNAs) can be faulty. Without a good sgRNA, Cas9 is ineffective and leaves the target gene intact: a false negative if the gene is actually essential.
Through the JGI’s Community Science Program, the scientists thus set out to identify which sgRNA sequences were reliable. Click here to read the science highlight on the JGI website.
- « Previous Page
- 1
- …
- 21
- 22
- 23
- 24
- 25
- …
- 46
- Next Page »
Was this page useful?