Within the archaeal domain, there is a group of tiny hitchhikers. These organisms are abundant and yet exceptionally small, with mini genomes to match. To fill in their nutrient gaps, they must latch onto a larger host — often, a fellow archaeal microbe. Recently, researchers used population genomics to find that while archaeal hitchhikers may often act as parasites, in other cases, they likely help their hosts.
JGI Extracts the Secrets of Secondary Metabolites
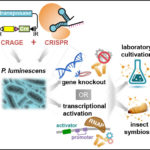
Microbial secondary metabolites, those molecules not essential for growth yet essential for survival, may now be easier to characterize following a JGI proof-of-concept study in which researchers paired CRISPR and CRAGE technologies. CRAGE (developed by a JGI team led by Yasuo Yoshikuni) offers CRISPR a point of entry into microbes that it previously lacked. Then, by using CRISPR to knock out or activate genes, researchers at the JGI were able to monitor loss- and gain-of-function, with the analytical data showing peaks and valleys in secondary metabolites as genes are edited. The pairing proved to rapidly confirm enhanced production of 22 metabolites from six biosynthetic gene clusters. One of those was a metabolite from a previously uncharacterized biosynthetic gene cluster. Learn more on the JGI website.
Congratulations to Biosciences Area Director’s Award Recipients
Numerous Biosciences Area personnel are among the 2021 Berkeley Lab Director’s Awards honorees. This annual program recognizes outstanding contributions by employees to all facets of Lab activities. A complete list of winners can be found here. The 10th annual Director’s Awards ceremony will take place on November 18 at noon.
Congratulations to Biosciences Area Director’s Award Recipients
Numerous Biosciences Area personnel are among the 2020 Berkeley Lab Director’s Awards honorees. This annual program recognizes outstanding contributions by employees to all facets of Lab activities. A complete list of winners can be found here. The ninth annual Director’s Awards ceremony will take place (virtually) on November 12 at 3 PM.
JGI Overhauls Perception of Inovirus Diversity
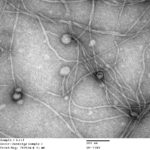
Inoviruses are filamentous viruses with small, single-stranded DNA genomes and a unique chronic infection cycle. In Nature Microbiology, a team led by DOE Joint Genome Institute (JGI) researchers applied machine learning to publicly available microbial genomes and metagenomes to search for inoviruses. The search tool combed through more than 70,000 microbial and metagenome datasets, ultimately identifying more than 10,000 inovirus-like sequences compared to the 56 previously known inovirus genomes. The results revealed inoviruses are in every major microbial habitat—including soil, water, and humans—around the world.
“We’re not sure why we systematically manage to miss them; maybe it’s due to the way we currently isolate and extract viruses,” said the study’s lead author Simon Roux, a JGI research scientist in the Environmental Genomics group. Click here to read the full story on the JGI site.
Was this page useful?