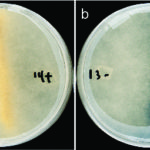
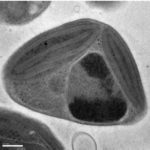
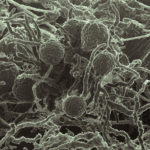
In heritable mutualisms, hosts pass on beneficial symbionts between generations. The origin of this relationship though, is often antagonistic and the parasite first needs to secure its own transmission before working with the host. Using the mutualistic relationship between the plant pathogenic fungus Rhizopus microsporus (Rm) and Burkholderia endobacteria, JGI and Cornell University researchers reported on how the antagonistic-to-mutualistic transition occurs in Nature Communications. The team found that the oil-producing fungus is highly dependent on the Burkholderia endobacteria to proliferate both sexually and asexually. This dependence is consistent with the addiction model of mutualism evolution. Read more on the JGI website.
JGI Comparative Genomics of Humongous Fungus Helps Explain Size, Pathogenicity
As part of an international team, researchers at the Joint Genome Institute (JGI) helped sequence and analyze the genomes of four fungi of the genus Armillaria. Often called the humongous fungus, Armillaria form some of the planet’s largest living organisms. They are also among the most devastating fungal pathogens, capable of breaking down all of the components of a host plant’s cell walls.
Tiny Green Algae Reveal Large Genomic Variation
A decade after the complete representative genomes of three Ostreococcus groups were sequenced, an international team of researchers including scientists at the Department of Energy Joint Genome Institute (DOE JGI) have resequenced and analyzed the genomes of 13 members of a natural population from the northwest Mediterranean Sea. The analysis revealed that the O. tauri population is larger than anticipated, with high genetic and phenotypic diversity that is influenced by the algae’s natural resistance to ocean viruses. Understanding the genetic variability of various Ostreococcus strains will help researchers understand how environmental changes affect their abundance and ability to photosynthesize. The work was reported in Science Advances. Read more in this DOE JGI Science Highlight.
Fungal Enzyme Clusters Promote Efficient Biomass Breakdown
Fungi, particularly those found in the digestive tracts of ruminant herbivores such as cattle, sheep, and goats, are very good at overcoming the resistance of plant cell walls to degradation—a major hurdle in the quest to produce sustainable fuels and chemicals from bioenergy feedstocks. Now, an international group of researchers has identified protein scaffolds in anaerobic gut fungi that provide docking sites for various enzymes, keeping them in place so that they can work together more effectively. As reported in the May 26 issue of Nature Microbiology, the structures are analogous to cellulosome complexes in anerobic bacteria, but this is the first time they have been found in fungi.
DOE JGI Finds A New Major Gene Expression Regulator in Fungi
Just four letters – A, C, T, and G – make up an organism’s genetic code. Changing a single letter, or base, can lead to changes in protein structures and functions, impacting an organism’s traits. In addition, though, subtler changes can and do happen, involving modifications of the DNA bases themselves. In the May 8, 2017 issue of Nature Genetics, a team led by scientists at the U.S. Department of Energy Joint Genome Institute (JGI), a DOE Office of Science User Facility, report the prevalence of modifications in the earliest branches of the fungal kingdom. Read more in the JGI press release.
- « Previous Page
- 1
- …
- 3
- 4
- 5
- 6
- Next Page »
Was this page useful?