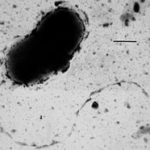
A high-throughput, cost effective method for discovering gene function in microbes under various environmental conditions is among the winners of R&D Magazine’s 2017 R&D 100 Award. The technology, called Dual Barcoded Shotgun Expression Library Sequencing (Dub-seq), was developed by Adam Arkin, Adam Deutschbauer, Vivek Mutalik, and Pavel Novichkov of the Environmental Genomics and Systems Biology (EGSB) Division. Dub-seq … Read more »
Dub-seq Named a Finalist for R&D 100 Awards
A method for discovering gene function in microbes developed by Adam Arkin, Adam Deutschbauer, Vivek Mutalik, and Pavel Novichkov of the Environmental Genomics and Systems Biology division has been shortlisted for R&D Magazine’s 2017 R&D 100 Award. The technology, called Dual Barcoded Shotgun Expression Library Sequencing (Dub-seq), combines the shotgun expression library method and next generation sequencing method developed through the ENIGMA program. Now in its 55th year, the prestigious R&D 100 Awards program recognizes the most innovative inventions of the prior year. The winners will be announced and honored at a black-tie ceremony to be held this fall in conjunction with the annual R&D 100 Conference.
ENIGMA Researchers Uncover Factors in Microbial Community Collapse
Microbial communities play key roles in all ecosystems and are essential to many industries and processes, such as agriculture and sewage treatment. Researchers in the Ecosystems and Networks Integrated with Molecular Assemblies (ENIGMA) Scientific Focus Area, a multi-institutional program that is managed by Lawrence Berkeley National Laboratory (Berkeley Lab), have developed a framework to quantify microbial community resilience in the face of environmental changes.
New Bacteria Groups, and Stunning Diversity, Discovered Underground
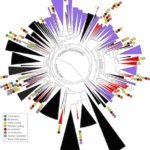
Lawrence Berkeley National Laboratory (Berkeley Lab) and UC Berkeley researchers have uncovered new clues about the roles of subsurface microbes in globally important cycles. Jill Banfield, senior faculty scientist in the Earth & Environmental Systems Area and professor at UC Berkeley, led the research team that studied soil and water samples containing subsurface microbes collected at a Colorado River basin field site. DNA sequencing of these microbes was performed at the Joint Genome Institute (JGI), a DOE Office of Science User Facility.
As reported online October 24 in the journal Nature Communications, the scientists netted genomes from 80 percent of all known bacterial phyla, a remarkable degree of biological diversity at one location. They also discovered 47 new phylum level bacterial groups, naming many of them after influential microbiologists and other scientists, including ten in the Biosciences Area (with the form Candidatus Surnamebacteria). Phyla-level names have been proposed for Molecular Biophysics & Integrated Bioimaging Division’s Cheryl Kerfeld, Krishna Niyogi, and Jennifer Doudna; Environmental Genomics & Systems Biology’s Louise Glass, Kathleen Ryan, Steven Brenner, Mary Wildermuth, and Judy Wall; and the JGI’s John Vogel and Tanja Woyke. The researchers analyzed the metabolic interactions of these and other subsurface microbes to better understand their roles in ecosystem resilience. Read the full story at the Berkeley Lab News Center.
Raising the Bar for Bacterial Interactions Networks
Scientists working as part of a multi-investigator ENIGMA (Ecosystems & Networks Integrated with Genes and Molecular Assemblies Scientific Focus Area) project have determined that protein-protein interactions occur more frequently among proteins with similar functions. The team of researchers, led by Gareth Butland in the Environmental Genomics & Systems Biology Division, used high throughput functional genomics to study the protein-protein interactome of the model sulfate reducing bacterium Desulfovibrio vulgaris Hildenborough. Their findings critically re-evaluate published bacterial interaction networks and establish benchmarks for high confidence protein interactomes. The manuscript can be found online at Molecular and Cellular Proteomics.
Was this page useful?