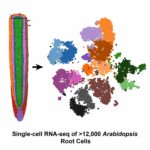
Researchers led by Diane Dickel have successfully adapted an open-source RNA analysis platform to study gene expression in individual plant cells. The method, called Drop-seq, was developed at Harvard Medical School in 2015 and had previously been used only in animal cells. Dickel and her colleagues at the DOE Joint Genome Institute (JGI) teamed up with researchers from UC Davis who had perfected a protoplasting technique for root tissue from Arabidopsis thaliana (mouse-ear cress). After preparing samples of more than 12,000 Arabidopsis root cells, the group was thrilled when the Drop-seq process went smoother than expected. Their results were published in Cell Reports.
JGI Maps Bacterial Genes That Regulate Microbe-Plant Root Interactions
A plant’s health and development is influenced by the complex community of microbes that surrounds it. Researchers at the Joint Genome Institute (JGI) and their collaborators at the Howard Hughes Medical Institute at the University of North Carolina have identified some 350 genes of the bacterium Pseudomonas simiae that positively or negatively impact how effectively this beneficial microbe colonizes plant roots. Cataloging these genes—and understanding the cellular functions that they’re involved in—is the first step toward developing targeted approaches to improving plant health and growth for a number of applications. The results of the study were published in PLOS Biology. Read more on the JGI website..
Was this page useful?