Sorghum bicolor (L.) Moench is an African grass that adroitly handles droughts, floods and poor soils. While sorghum is drought-tolerant, the crop’s precise response is dependent on when exactly water becomes a limiting factor – before or after flowering. Reported in PNAS the week of December 2, 2019, is the first paper that describes sorghum’s response to drought, from a large-scale field experiment led by a multi-institutional consortium to uncover the mechanisms behind sorghum’s capacity to produce high yields despite drought conditions. Read the full highlight on the JGI website.
JGI Overhauls Perception of Inovirus Diversity
Inoviruses are filamentous viruses with small, single-stranded DNA genomes and a unique chronic infection cycle. In Nature Microbiology, a team led by DOE Joint Genome Institute (JGI) researchers applied machine learning to publicly available microbial genomes and metagenomes to search for inoviruses. The search tool combed through more than 70,000 microbial and metagenome datasets, ultimately identifying more than 10,000 inovirus-like sequences compared to the 56 previously known inovirus genomes. The results revealed inoviruses are in every major microbial habitat—including soil, water, and humans—around the world.
“We’re not sure why we systematically manage to miss them; maybe it’s due to the way we currently isolate and extract viruses,” said the study’s lead author Simon Roux, a JGI research scientist in the Environmental Genomics group. Click here to read the full story on the JGI site.
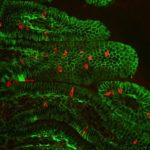
Epic Research Endeavor Reveals Cause of Deadly Digestive Disease in Children
Berkeley Lab geneticist Len Pennacchio and his team helped a group of Israeli clinical researchers solve the mystery of a rare inherited disease that causes extreme, sometimes fatal, chronic diarrhea in children. The nearly decade-long investigation not only led to the discovery of a novel protein-coding gene that is critical for intestinal function, but also expanded our understanding of regulatory sequences in the human genome. The results were recently published in Nature.
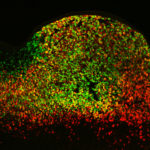
Gene Enhancers Are Important Despite Apparent Redundancy
Though once thought to be “junk DNA,” enhancers are extremely important, regulating the expression of specific genes that ultimately determine a cell’s properties and functions. However, there are many more enhancers than genes and their relationship is unclear. A team of scientists in the Environmental Genomics & Systems Biology Division have turned their attention to this relationship and the overall importance of enhancers to development. By better linking the genomic complement of an organism with its expressed characteristics, their work offers new insights that further the growing field of systems biology, which seeks to gain a predictive understanding of living systems.
JGI Maps Bacterial Genes That Regulate Microbe-Plant Root Interactions
A plant’s health and development is influenced by the complex community of microbes that surrounds it. Researchers at the Joint Genome Institute (JGI) and their collaborators at the Howard Hughes Medical Institute at the University of North Carolina have identified some 350 genes of the bacterium Pseudomonas simiae that positively or negatively impact how effectively this beneficial microbe colonizes plant roots. Cataloging these genes—and understanding the cellular functions that they’re involved in—is the first step toward developing targeted approaches to improving plant health and growth for a number of applications. The results of the study were published in PLOS Biology. Read more on the JGI website..
Was this page useful?