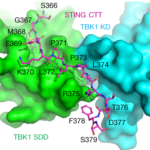
The crystallographic study of STING (stimulator of interferon genes), a transmembrane protein that plays a key role in innate immunity, in complex with TBK1 (serine/threonine-protein kinase), an enzyme that regulates the inflammatory response to foreign DNA, is extremely challenging due to weakly diffracting crystals. But thanks to the expertise of Berkeley Center for Structural Biology (BCSB) scientists, researchers from Texas A&M University (TAMU) were able to pinpoint the conserved motif of STING that mediates the recruitment and activation of TBK1. They published their results in Nature.
Researchers Create Comprehensive Model of Transcription Preinitiation Complex
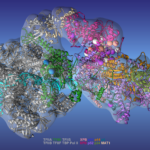
Researchers led by Ivaylo Ivanov of Georgia State University have produced a comprehensive model of the human transcription preinitiation complex (PIC), a vital assembly of proteins responsible for regulating gene expression. The new model is the most complete to date and provides mechanistic insights into how mutations affecting one component—human transcription initiation factor IIH, or TFIIH—lead to three inherited genetic diseases. Berkeley Lab Biosciences’ Susan Tsutakawa, a research scientist in Molecular Biophysics and Integrated Bioimaging (MBIB), and John Tainer, a professor at the University of Texas MD Anderson Cancer Center and visiting faculty in MBIB, were part of the team. Results from this work were recently published in Nature Structural & Molecular Biology.
Technique for Studying Gene Expression Takes Root in Plants
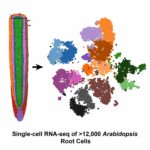
Researchers led by Diane Dickel have successfully adapted an open-source RNA analysis platform to study gene expression in individual plant cells. The method, called Drop-seq, was developed at Harvard Medical School in 2015 and had previously been used only in animal cells. Dickel and her colleagues at the DOE Joint Genome Institute (JGI) teamed up with researchers from UC Davis who had perfected a protoplasting technique for root tissue from Arabidopsis thaliana (mouse-ear cress). After preparing samples of more than 12,000 Arabidopsis root cells, the group was thrilled when the Drop-seq process went smoother than expected. Their results were published in Cell Reports.
JGI Researchers Featured in mSystems Special Issue
Three Joint Genome Institute researchers are among the authors who offered perspectives on what the next five years of innovation could look like for a special issue of the journal mSystems. In one article, Micro-Scale Applications head Rex Malmstrom and Metagenome Program head Emiley Eloe-Fadrosh outline more targeted approaches to reconstruct individual microbes in an environmental sample. In a separate article, research scientist Simon Roux, a member of Eloe-Fadrosh’s Environmental Genomics group, makes a pitch for readers to get involved in the developing field of virus ecogenomics.
Read more from JGI.
Superfacility Framework Advances Photosynthesis Research
An article published in the Computing Sciences News Center describes how Biosciences researchers are using a superfacility framework of experimental instrumentation with computational and data facilities to unravel the long-standing mystery of how Photosystem (PSII) works. The protein complex plays a crucial role in photosynthesis, making it key to achieving artificial photosynthesis that could produce fuels using sunlight and carbon dioxide. Researchers—led by Vittal Yachandra, Junko Yano, and Jan Kern in Molecular Biophysics and Integrated Bioimaging (MBIB)—recently began using ESnet to enable real-time processing of experimental data collected at the SLAC National Accelerator Laboratory’s Linac Coherent Light Source (LCLS) at NERSC to observe this water-splitting protein in action. Asmit Bhowmick, a postdoctoral researcher in the laboratory of MBIB senior scientist Nicholas Sauter, is quoted in the article.
- « Previous Page
- 1
- …
- 95
- 96
- 97
- 98
- 99
- …
- 213
- Next Page »
Was this page useful?