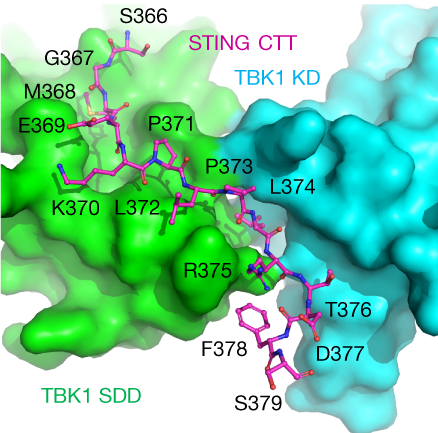
The C-terminal tail (CTT) of STING, shown by the magenta ball-and-stick model, binds to the interface between the kinase domain (cyan) and the scaffold and dimerization domain (green) of a TBK1 dimer.
The crystallographic study of STING (stimulator of interferon genes), a transmembrane protein that plays a key role in innate immunity, in complex with TBK1 (serine/threonine-protein kinase), an enzyme that regulates the inflammatory response to foreign DNA, is extremely challenging due to weakly diffracting crystals. But thanks to the expertise of Berkeley Center for Structural Biology (BCSB) scientists, researchers from Texas A&M University (TAMU) were able to pinpoint the conserved motif of STING that mediates the recruitment and activation of TBK1.
The TAMU team, led by Pingwei Li, published their results in Nature. Banumathi Sankaran, a research scientist in the BSCB at the Advanced Light Source (ALS), was an author on the paper. Sankaran leads the Collaborative Crystallography (CC) mail-in service at the ALS, which is run under the aegis of the ALS-ENABLE program. She collected X-ray crystallography data at BCSB beamlines 8.2.2 and 5.0.2 that enabled the researchers to obtain 3.2 to 3.4 angstrom resolution structures for both the STING C-terminal peptide and the C-terminal ligand binding domain bound to TBK1.
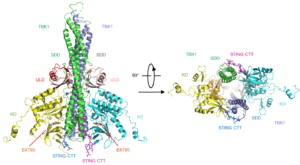
Mouse TBK1 in complex with the human STING CTT. Kinase domains (KD) are in yellow and cyan; ubiquitin-like domains (ULD) are in pink and red; scaffold and dimerization domains (SDD) are in green and slate; and the CTT of STING is shown by the blue and magenta ball-and-stick models.
Due to the relatively low resolution of the diffraction data, modeling the peptide bound to TBK1 was also challenging. Sankaran helped collect data from several Se-Met derivative crystals, which crucially helped confirm the sequence and validate the initial model. The TAMU researchers were then able to determine the experimental phases by single wavelength anomalous diffraction (SAD) and calculate an anomalous difference map to nail down the Se sites using the Phenix software suite, developed under the guidance of Paul Adams, Director of the Molecular Biophysics and Integrated Bioimaging (MBIB) Division.