Jennifer Doudna, faculty scientist in the Molecular Biophysics & Integrated Bioimaging Division, UC Berkeley professor of chemistry and of molecular and cell biology, and Howard Hughes Medical Institute investigator, will receive the 2018 National Academy of Sciences (NAS) Award in Chemical Sciences. According to the NAS award announcement, Doudna is honored for her “pioneering discoveries on how RNA can fold to function in complex ways,” and her invention, with Emmanuelle Charpentier, of “the technology for efficient site-specific genome engineering using the CRISPR/Cas9 nucleases for genome editing — a breakthrough technology which has had an immediate and wide impact on all areas of both basic and applied life sciences.”
‘Magic Pools’ Approach Accelerates Study of Novel Bacteria
To characterize the functions of genes in newly discovered bacteria, microbiologists introduce mutations using mobile DNA snippets called transposons delivered via a vector. But finding an effective transposon mutagenesis system for a novel microorganism through trial and error can be a time-consuming and expensive endeavor.
To streamline the process, Environmental Genomics and Systems Biology (EGSB) Division researchers, led by Adam Deutschbauer and Adam Arkin, have developed a new technique to test hundreds of different transposon vector variants in parallel within what the investigators term “magic pools.”
The approach was detailed in mSystems, an open access journal from the American Society for Microbiology.
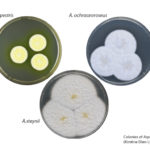
JGI, JBEI Present First Results of Genus-Wide Aspergillus Project
Found in microbial communities around the world, Aspergillus fungi are pathogens, decomposers, and important sources of biotechnologically-important enzymes. In a study published ahead the week of January 8, 2018 in the Proceedings of the National Academy of Sciences, a team led by researchers at the Technical University of Denmark (DTU), the Joint Genome Institute (JGI), and the DOE’s Joint BioEnergy Institute (JBEI) report the first results of a long-term plan to sequence, annotate and analyze the genomes of 300 Aspergillus fungi. These findings are a proof of concept of novel methods to functionally annotate genomes in order to more quickly identify genes of interest. Read more on the JGI website.
JGI Helps Analyze Dry Rot’s Adaptation to Built Environments
Unlike white rots, brown rots break down only the cellulose and hemicellulose, leaving the lignin behind. The brown rot Serpula lacrymans is typically found in spruce and other conifers in boreal forests. As these trees were harvested for constructing buildings, the dry rot fungus migrated indoors and across borders, adapting to thrive in manmade environments. Reported January 5, 2018 in The ISME Journal, a comparative genomics analysis by a team led by University of Oslo scientists and including JGI researchers lends insights on how the fungus has responded to manmade changes in its ecological habitat, adapting to thrive in built environments. Learn more on the JGI website.
Microbial Metabolism in Real World Native Biocrusts
Biological soil crusts, or biocrusts, contain communities of microorganisms—including fungi, bacteria, and archaea—that dwell together within the uppermost millimeters of soil in arid lands. These microbes can exist for extended periods in a desiccated, dormant state, becoming metabolically active when it rains. Understanding how biocrust microbial communities adapt to their harsh environments could help shed light on the roles of soil microbes in the global carbon cycle. Berkeley Lab scientists led by Trent Northen’s group in Environmental Genomics and Systems Biology (EGSB) found that specific compounds are transformed by and strongly associated with specific bacteria in native biological soil crust. The researchers reported their findings in a paper published January 2 in Nature Communications.
- « Previous Page
- 1
- …
- 133
- 134
- 135
- 136
- 137
- …
- 214
- Next Page »
Was this page useful?