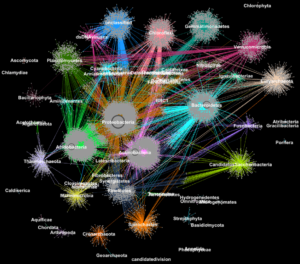
On a social network like Facebook, each user (person or organization) is represented as a node and the connections (relationships and interactions) between them are called edges. By analyzing these connections, researchers can learn a lot about each user. In biology, similar graph-clustering algorithms can be used to understand the proteins that perform most of life’s functions. Today, advanced high-throughput technologies allow researchers to capture hundreds of millions of proteins, genes and other cellular components at once and in a range of environmental conditions. Clustering algorithms are then applied to these datasets to identify patterns and relationships that may point to structural and functional similarities. Though these techniques have been widely used for more than a decade, they cannot keep up with the torrent of biological data being generated by next-generation sequencers and microarrays. In fact, very few existing algorithms can cluster a biological network containing millions of nodes (proteins) and edges (connections). That’s why a team of Berkeley Lab researchers, including scientists at the Joint Genome Institute (JGI), took one of the most popular clustering approaches in modern biology—the Markov Clustering (MCL) algorithm—and modified it to run quickly, efficiently and at scale on distributed-memory supercomputers. In a test case, their high-performance algorithm—called HipMCL—achieved a previously impossible feat: clustering a large biological network containing about 70 million nodes and 68 billion edges in a couple of hours, using approximately 140,000 processor cores on the National Energy Research Scientific Computing Center’s (NERSC) Cori supercomputer. A paper describing this work was recently published in the journal Nucleic Acids Research. Read the whole story on the Berkeley Lab Computational Research Division site.



