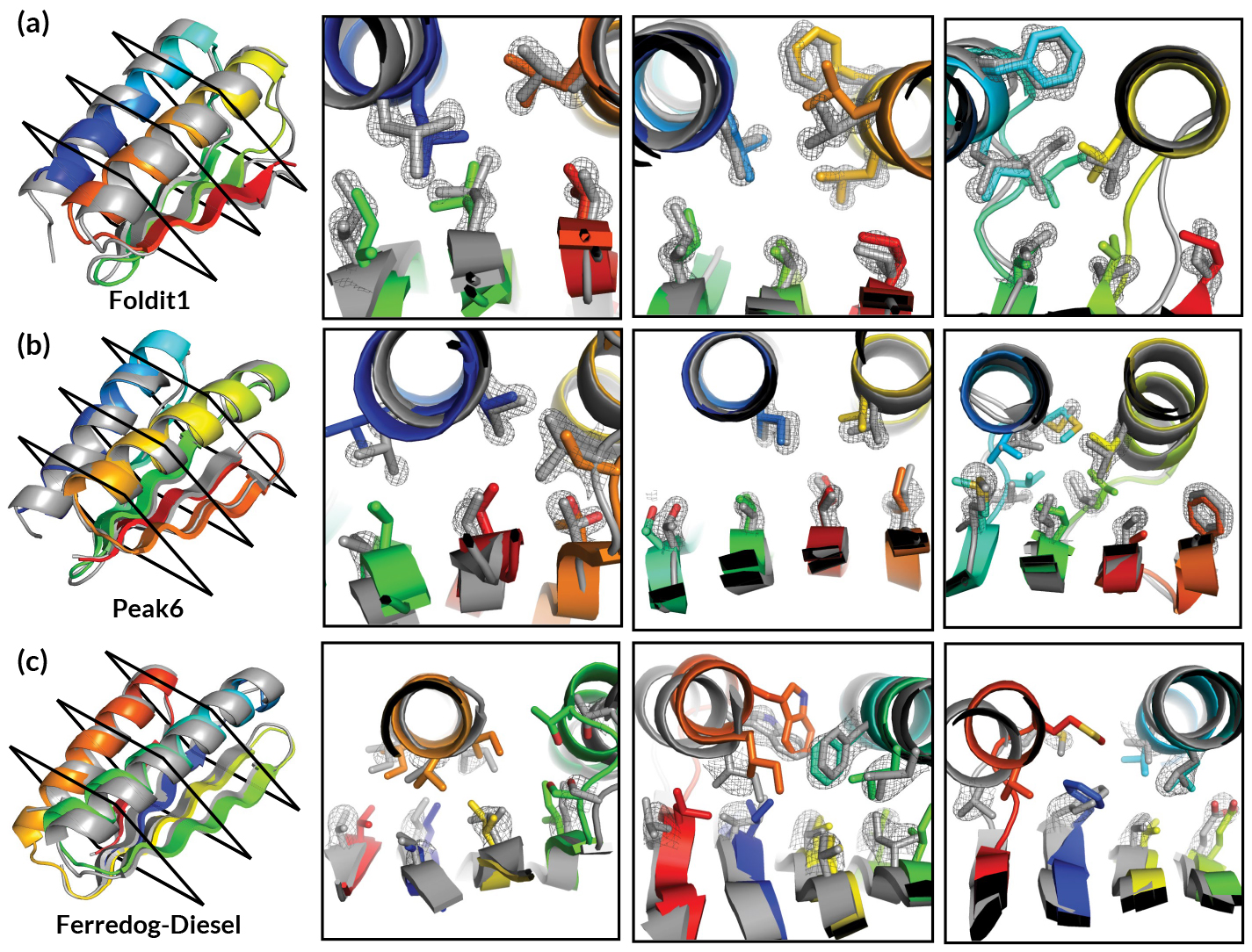
Berkeley Center for Structural Biology (BCSB) beamlines 5.0.2, 8.2.1, and 8.2.2 at the Advanced Light Source (ALS) were used to experimentally verify de novo protein structures designed by citizen scientists playing the computer game Foldit.
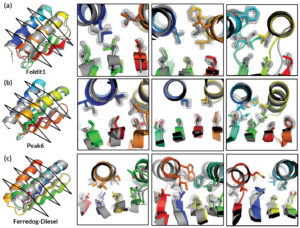
Comparison of high-resolution x-ray crystallography results (gray) with Foldit player-designed protein models (rainbow).
First introduced in 2008, Foldit is a free online game developed by University of Washington researchers to crowdsource problems in protein modeling. Gameplay incorporates principles of protein folding by giving higher scores for things like removing voids, covering hydrophobic strands, and clearing clashes between amino-acid side chains. The game has been refined over time; for example, in the past Foldit was primarily applied to the prediction or modification of known protein structures.
However, for a recent study, researchers presented players with a far more expansive task: given a completely unfolded amino-acid chain, craft a de novo protein structure. The challenge resulted in 146 designs, 56 of which were found to adopt stable structures when produced in a lab. Four of the structures were verified experimentally (three at the ALS).
The Foldit designs—crafted by players with little or no biochemical training—turned out to be as good as those generated by experts or computers, with an equivalent success rate in the lab and similar folding stabilities. Moreover, the designs were more structurally diverse because players were willing to explore whole new regions in protein space.
For more, read the ALS Science Highlight.