Through a decade-long time-series study recently reported in Microbiome, a team led by Rick Cavicchioli of Australia’s University of New South Wales and including JGI researchers found that light-harvesting bacteria in Antarctica’s Ace Lake defy the norm when it comes to nutrient cycling in polar regions. While microbial populations in other Antarctic lakes and the nearby Southern Ocean shift from phototrophs to archaea when sunlight becomes scant, Ace Lake’s bacteria instead go through a boom and bust cycle aligned with light availability. Read more on the JGI website.
Decoding Messages in the Body’s Microscopic Metropolises
A study aimed at identifying and examining the small messenger proteins used by microbes living on and inside humans has revealed an astounding diversity of more than 4,000 families of molecules – many of which have never been described previously. The research, led by Stanford University and now published in Cell, lays the groundwork for future investigations into how the trillions of bacteria, archaea, and fungi that compose human microbiomes compete for resources, attack and co-exist with one another, and interact with our own cells.
JGI Overhauls Perception of Inovirus Diversity
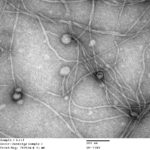
Inoviruses are filamentous viruses with small, single-stranded DNA genomes and a unique chronic infection cycle. In Nature Microbiology, a team led by DOE Joint Genome Institute (JGI) researchers applied machine learning to publicly available microbial genomes and metagenomes to search for inoviruses. The search tool combed through more than 70,000 microbial and metagenome datasets, ultimately identifying more than 10,000 inovirus-like sequences compared to the 56 previously known inovirus genomes. The results revealed inoviruses are in every major microbial habitat—including soil, water, and humans—around the world.
“We’re not sure why we systematically manage to miss them; maybe it’s due to the way we currently isolate and extract viruses,” said the study’s lead author Simon Roux, a JGI research scientist in the Environmental Genomics group. Click here to read the full story on the JGI site.
Updating the Genome Data Sharing Agreement
Nearly 20 years ago in Fort Lauderdale, Fla., genome data producers and data users came to an accord on the use of genome sequencing data released to the public domain. In particular, they agreed that the data was freely available for use and access by the scientific community before those data are used for publication. The Fort Lauderdale Agreement did not include defined policies on data usage, and has led to years of debate, such as whether or not there was a tacit acknowledgement that data generators would have the right of first publication on the data they produced and freely shared.
In a policy paper published January 25, 2019 in Science, 50 coauthors, with 54 unique affiliations from 18 countries, call for a “clear policy that protects public data from restrictions.” The international consortium includes Nikos Kyrpides of the Biosciences Area’s Environmental Genomics & Systems Biology (EGSB) Division at Lawrence Berkeley National Laboratory (Berkeley Lab).
JGI Helps Develop Metagenomic Clustering Algorithm Powered by HPC
On a social network like Facebook, each user (person or organization) is represented as a node and the connections (relationships and interactions) between them are called edges. By analyzing these connections, researchers can learn a lot about each user. In biology, similar graph-clustering algorithms can be used to understand the proteins that perform most of life’s functions. Today, advanced high-throughput technologies allow researchers to capture hundreds of millions of proteins, genes and other cellular components at once and in a range of environmental conditions. Clustering algorithms are then applied to these datasets to identify patterns and relationships that may point to structural and functional similarities. Though these techniques have been widely used for more than a decade, they cannot keep up with the torrent of biological data being generated by next-generation sequencers and microarrays. In fact, very few existing algorithms can cluster a biological network containing millions of nodes (proteins) and edges (connections). That’s why a team of Berkeley Lab researchers, including scientists at the Joint Genome Institute (JGI), took one of the most popular clustering approaches in modern biology—the Markov Clustering (MCL) algorithm—and modified it to run quickly, efficiently and at scale on distributed-memory supercomputers. In a test case, their high-performance algorithm—called HipMCL—achieved a previously impossible feat: clustering a large biological network containing about 70 million nodes and 68 billion edges in a couple of hours, using approximately 140,000 processor cores on the National Energy Research Scientific Computing Center’s (NERSC) Cori supercomputer. A paper describing this work was recently published in the journal Nucleic Acids Research. Read the whole story on the Berkeley Lab Computational Research Division site.
Was this page useful?