Berkeley scientists — including Biosciences’s Jillian Banfield (secondary affiliation with the Environmental Genomics & System Biology Division) and Jennifer Doudna (Molecular Biophysics & Integrated Bioimaging Division) — have discovered simple CRISPR systems similar to CRISPR-Cas9 — a gene-editing tool that has revolutionized biology — in previously unexplored bacteria that have eluded efforts to grow them in the laboratory. Read more at Berkeley News.
Berkeley scientists — including Biosciences’s Jillian Banfield (secondary affiliation with the Environmental Genomics & System Biology Division) and Jennifer Doudna (Molecular Biophysics & Integrated Bioimaging Division) — have discovered simple CRISPR systems similar to CRISPR-Cas9 — a gene-editing tool that has revolutionized biology — in previously unexplored bacteria that have eluded efforts to grow them in the laboratory. Read more at Berkeley News.
New Bacteria Groups, and Stunning Diversity, Discovered Underground
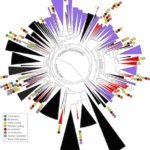
Lawrence Berkeley National Laboratory (Berkeley Lab) and UC Berkeley researchers have uncovered new clues about the roles of subsurface microbes in globally important cycles. Jill Banfield, senior faculty scientist in the Earth & Environmental Systems Area and professor at UC Berkeley, led the research team that studied soil and water samples containing subsurface microbes collected at a Colorado River basin field site. DNA sequencing of these microbes was performed at the Joint Genome Institute (JGI), a DOE Office of Science User Facility.
As reported online October 24 in the journal Nature Communications, the scientists netted genomes from 80 percent of all known bacterial phyla, a remarkable degree of biological diversity at one location. They also discovered 47 new phylum level bacterial groups, naming many of them after influential microbiologists and other scientists, including ten in the Biosciences Area (with the form Candidatus Surnamebacteria). Phyla-level names have been proposed for Molecular Biophysics & Integrated Bioimaging Division’s Cheryl Kerfeld, Krishna Niyogi, and Jennifer Doudna; Environmental Genomics & Systems Biology’s Louise Glass, Kathleen Ryan, Steven Brenner, Mary Wildermuth, and Judy Wall; and the JGI’s John Vogel and Tanja Woyke. The researchers analyzed the metabolic interactions of these and other subsurface microbes to better understand their roles in ecosystem resilience. Read the full story at the Berkeley Lab News Center.
Was this page useful?