The supply of a plant-derived anti-cancer drug can finally meet global demand after a team of scientists from Denmark and the U.S. engineered yeast to produce the precursor molecules, which could previously only be obtained in trace concentrations in the native plant. A study describing the breakthrough was recently published in Nature. The international team included four researchers from the Biological Systems and Engineering Division: Leanne Jade G. Chan, Edward Baidoo, Christopher J. Petzold, and Jay D. Keasling.
New Protein Functions from Beneficial Human Gut Bacterium
Researchers in the Environmental Genomics and Systems Biology (EGSB) and Biological Systems and Engineering (BSE) Divisions at Berkeley Lab employed a large-scale functional genomics approach to systematically characterize Bacteroides thetaiotaomicron, a beneficial bacterium prevalent in the human gut. They performed hundreds of genome-wide fitness assays and identified new functions for 40 proteins, including antibiotic tolerance, polysaccharide degradation, and colonization of the GI tract in germ-free mice.
Get a Move On: Protein Translates Chemistry into Motion
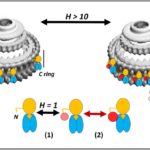
The protein CheY plays a role in relaying sensory signals from chemoreceptors to the rotary motor at the base of the tail-like appendage, or flagellum, that protrudes from the cell body of certain bacteria and eukaryotic cells. It has been studied as a model for dissecting the mechanism of allostery—the process by which the binding of biological macromolecules (mainly proteins) at one location regulates activity at another, often distant, functional site. When it is transiently phosphorylated in response to chemotactic cues, CheY’s binding affinity for a flagellar motor switch protein called FliM is enhanced. CheY binding to FliM changes the direction of flagellar rotation from counterclockwise to clockwise.
Using X-ray footprinting with mass spectroscopy (XFMS), a team led by Shahid Khan, a senior scientist with the Molecular Biology Consortium, established that CheY changes shape when it tethers to the motor, and further parsed the contribution of phosphorylation to this shape change. The results of the XFMS experiments validated atomistic molecular dynamics (MD) predictions of the architecture of the allosteric communication network, marking the first time that XFMS has been used to validate protein dynamics simulations at single-residue resolution sampled over the complete protein.
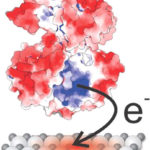
X-ray Footprinting Reveals Secrets of ‘Metal-Breathing’ Bacterium
A team of Berkeley Lab researchers conducted X-ray footprinting mass spectrometry (XFMS) experiments at the Lab’s Advanced Light Source (ALS) to pinpoint how a protein of the bacterium Shewanella oneidensis transfers electrons to a metal oxide substrate. The research was led by Caroline Ajo-Franklin, whose lab is part of the Molecular Foundry and who holds a secondary appointment in the Molecular Biophysics and Integrated Bioimaging (MBIB) division, in collaboration with Corie Ralston, also of MBIB. Tatsuya Fukushima, a former postdoc in Ajo-Franklin’s lab, and Sayan Gupta, a member of Ralston’s lab, were co-first authors on the paper published in the Journal of the American Chemical Society. The study, which identified an unexpectedly small and weak binding site, also benefitted from expertise and tools contributed by Joint BioEnergy Institute (JBEI) and Biological Systems and Engineering (BSE) researchers Christopher Petzold and Leanne Jade Chan. Read more at the Berkeley Lab News Center.
JBEI/BSE Paper Among PLOS ONE Top 10% Most Cited Articles
 A paper by Joint BioEnergy Institute (JBEI) and Biological Systems and Engineering (BSE) researchers has been ranked among the top 10% most cited PLOS ONE articles. “A Thermophilic Ionic liquid-tolerant Cellulase Cocktail for the Production of Cellulosic Biofuels” published in 2012 has already been viewed 8,871 times and cited 50 times as of today. The paper reports the development of an Ionic Liquid-tolerant cellulase cocktail by combining thermophilic bacterial glycoside hydrolases produced by a mixed consortia with recombinant glycoside hydrolases.
A paper by Joint BioEnergy Institute (JBEI) and Biological Systems and Engineering (BSE) researchers has been ranked among the top 10% most cited PLOS ONE articles. “A Thermophilic Ionic liquid-tolerant Cellulase Cocktail for the Production of Cellulosic Biofuels” published in 2012 has already been viewed 8,871 times and cited 50 times as of today. The paper reports the development of an Ionic Liquid-tolerant cellulase cocktail by combining thermophilic bacterial glycoside hydrolases produced by a mixed consortia with recombinant glycoside hydrolases.
Was this page useful?