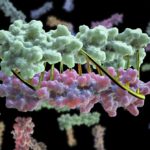
Bioscientists at the Advanced Light Source (ALS) at Berkeley Lab lent their expertise to a project led by scientists at the University of Washington to design proteins in the lab that zip together like DNA. The technique could enable the design of protein nanomachines to help diagnose and treat disease, allow for more precise engineering of cells, and perform a variety of other tasks.
Crystallization Screen Created by Berkeley Lab Biosciences Scientists Reaches the Market
 X-ray crystallography has been the most successful technique used to solve macromolecular structures, contributing several thousand new entries to the Protein Data Bank (PDB) every year. The protein crystal is the critical starting point for X-ray data collection, and consequently, its properties are correlated with the quality of the data and the level of detail that can be extracted for a macromolecular structure. However, proteins require solutions of specific composition to form crystals for structure determination studies. These specifications are usually determined from exposing the protein to several different solutions in a crystallization screen.
X-ray crystallography has been the most successful technique used to solve macromolecular structures, contributing several thousand new entries to the Protein Data Bank (PDB) every year. The protein crystal is the critical starting point for X-ray data collection, and consequently, its properties are correlated with the quality of the data and the level of detail that can be extracted for a macromolecular structure. However, proteins require solutions of specific composition to form crystals for structure determination studies. These specifications are usually determined from exposing the protein to several different solutions in a crystallization screen.
A team of researchers in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division led by Paul Adams and Jose Henrique Pereira have developed a new crystal screen, the Berkeley Screen, with 96 conditions proven to be highly effective at producing crystals for structural determination. The Berkeley Screen is now available to the wider crystallography community commercially.
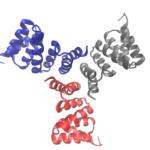
Designing Cyclic Oligomers: Greater than the Sum of Their Parts
Cyclic proteins that assemble from multiple identical subunits (homo-oligomers) play key roles in many biological processes, including enzymatic catalysis and function and cell signaling. Researchers in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division worked with University of Washington’s David Baker, who led a team to design in silico and crystallize self-assembling cyclic homo-oligomer proteins.
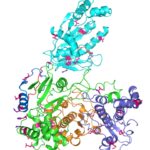
Researchers Gain Insight into Protein Critical to Zika Virus Reproduction
Zika virus is a mosquito-borne infectious disease linked to certain birth defects in infants in South and Central America and the United States. A Lawrence Berkeley National Laboratory (Berkeley Lab) researcher, Banumathi Sankaran, worked as part of a multi-institutional team led by Cheng Kao, professor at Indiana University, and Pingwei Li, associate professor at Texas A&M University (TAMU), to map a key viral protein called NS5. Necessary to virus reproduction, NS5 contains two enzyme activities: one reduces the body’s ability to mount an immune response against infection and the other helps start the genetic replication process.
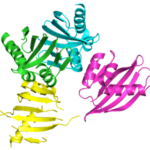
Designing Protein Cavities from Curved Beta Sheets
Curved beta sheets are important for the architecture of protein cavities, such as enzyme active sites and ligand-binding pockets. Beginning by analyzing classic protein formations and running folding simulations, University of Washington (UW) researchers under the leadership of David Baker designed six protein folds inspired by naturally occurring protein superfamilies. A research report published in the January 13 issue of Science describes how a multi-institutional team of scientists compared the predicted models to physical structures of these designed proteins.
Was this page useful?