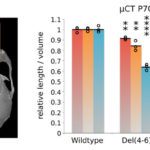
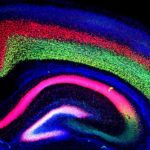
Biosciences researchers Axel Visel (JGI/EGSB) and Marco Osterwalder (EGSB) contributed to a study, led by researchers at the Max Planck Institute for Molecular Genetics, in which targeted genome editing and a transgenic reporter assay were used to characterize elements regulating Ihh (encoding Indian hedgehog) in mice. Indian hedgehog is a mammalian signaling protein involved with the development and proliferation of cells in cartilage. In humans, copy number variants (CNVs) upstream of Ihh cause localized phenotypes including premature fusion of the sutures of the skull and malformation of the phalanges. The study, published in Nature Genetics, showed that in mice Ihh is regulated by modular ensembles of enhancers (with individual tissue specificities) that appear to act in an additive manner. Despite apparent redundancy and overlapping function of enhancers, these ensembles—in which the correct number of each enhancer is present—are necessary for precise spatiotemporal control of developmental gene expression.
Researchers Collaborate to Create, Characterize Mouse Model of Autism
Using a genetic mouse model developed at Berkeley Lab with CRISPR/Cas9 editing technology, University of California, Davis, researchers led by Alexander Nord have examined the developmental impact of a specific mutation found in some rare cases of autism spectrum disorder (ASD). The project began while Nord—now an assistant professor with the Center for Neuroscience at UC Davis—was a postdoc in the Mammalian Functional Genomics Laboratory of Berkeley Lab co-authors Axel Visel, Len Pennacchio, and Diane Dickel. In a paper published June 26 in the journal Nature Neuroscience, the researchers report that mice with a loss-of-function mutation in one copy of the CHD8 gene have increased brain volume and cognitive impairment, similar to that seen in people with the same mutation. CHD8 encodes a protein responsible for packaging DNA in cells, which in turn controls gene expression during development. Read more from UC Davis.
JGI Researchers Release 1000+ Microbial Reference Genomes
Department of Energy Joint Genome Institute (DOE JGI) and Biosciences Environmental Genomics and Systems Biology (EGSB) division researchers have released 1,003 reference genomes for diverse bacteria and archea isolated from environments ranging from sea water and soil, to plants, and to cow rumen and termite guts. The release is the largest to date from JGI’s Genomic Encyclopedia of Bacteria and Archaea (GEBA) initiative, which seeks to fill in unexplored branches of the tree of microbial life. JGI’s Supratim Mukherjee and Rekha Seshadri were co-first authors on the paper published in Nature Biotechnology; senior author Nikos Kyrpides and co-authors Natalia Ivanova, Axel Visel, Tanja Woyke, and Yasuo Yoshikuni have secondary affiliations with EGSB. The genomes are publicly available through the Integrated Microbial Genomes with Microbiomes (IMG/M) system. Read more on the JGI website.
Finding Our Way Around DNA
Biosciences researchers collaborated with a team at the Salk Institute that developed a computational algorithm that integrates two different data types to make locating key regions within the genome more precise and accurate than other tools. The team’s method, recently described in Proceedings of the National Academy of Sciences, could help researchers conduct vastly more targeted searches for disease-causing genetic variants in the human genome, such as ones that promote cancer or cause metabolic disorders. Joseph Ecker, a Howard Hughes Medical Institute investigator and director of Salk’s Genomic Analysis Laboratory, was senior author of the study. Diane Dickel, Axel Visel and Len Pennacchio of the Environmental Genomics & Systems Biology Division, were co-authors, along with other researchers at the Salk Institute, UC San Diego, and UC San Francisco. Read more about the study in the Salk Institute press release.
Berkeley Lab Gets $4.6M in Functional Genomics Catalog Project
The Department of Energy’s Lawrence Berkeley National Laboratory (Berkeley Lab) is set to receive close to $4.6 million over four years as part of an ongoing federally funded project to create a comprehensive catalog for fundamental genomics research. This work will be part of the National Human Genome Research Institute’s Encyclopedia of DNA Elements ENCODE project.
The new Berkeley Lab grant, awarded at more than $1.1 million per year, will be used to establish the Center for In Vivo Characterization of ENCODE Elements (CIViC). It will be one of five characterization centers tasked with investigating how genomic elements function in vivo. CIViC will be led by principal investigators Len Pennacchio and Axel Visel, senior scientists at Berkeley Lab’s Environmental Genomics and Systems Biology Division. Research scientist Diane Dickel will be the center’s project manager. Read more in the Berkeley Lab News Center.
- « Previous Page
- 1
- …
- 3
- 4
- 5
- 6
- Next Page »
Was this page useful?