Frederik Schulz
Biologist Staff Scientist

Building: 91, Room 0460B5
Mail Stop: 91R0183
Phone: (510) 495-8559
fschulz@lbl.gov
https://jgi.doe.gov/who-we-are/frederik-schulz
https://github.com/NeLLi-team
Links
Divisions
- Science Programs
Secondary Affiliation:
Environmental Genomics and Systems Biology
- Comparative and Functional Genomics
Biography
Frederik Schulz joined the DOE Joint Genome Institute in 2016 and is leading the New Lineages of Life Group. His research is focussed on the discovery of novel bacterial, archaeal and eukaryotic microbes and viruses in environmental sequence data. Schulz employs multi-omics (metagenomics, metatranscriptomics, single cell genomics and phylogenomics) and machine learning to find novel functions that may impact microbiome structure and biogeochemical cycles. He provides his expertise to the JGI user community and supports various user projects in the Microbial Program. In an LDRD-funded project, Schulz established a single-cell-based discovery pipeline and enrichment metagenomics for terrestrial protists. These approaches enable novel insights into the diversity and important ecological roles of microeukaryotes in terrestrial ecosystems. Additionally, Schulz is leading a DTRA-funded program on agnostic pathogen detection and surveillance in complex samples.
Research Interests
-
Microbial symbiosis
Giant viruses and other eukaryotic viruses
Protists
Pathogen agnostic detection
Strain-resolved sequencing
Programs & Initiatives
Recent Publications
Related News
Congratulations to Biosciences Area Director’s Award Recipients
Several Biosciences Area personnel are among the 2024 recipients of Berkeley Lab Director’s Achievement Awards. The program recognizes outstanding contributions by employees to all aspects of Lab activities.
A Tool to Find Nomadic Genes that Help Microbes Adapt
Researchers at the JGI develop a tool to quickly and accurately identify mobile genetic elements like plasmids and viruses.
Biosciences Area FY23 LDRD Projects
The projects of 22 Biosciences Area scientists and engineers received funding through the FY23 Laboratory Directed Research and Development (LDRD) program.
Recent Publications
Divisions
- Science Programs
Secondary Affiliation:
Environmental Genomics and Systems Biology
- Comparative and Functional Genomics
Research Interests
Dr. Li Lei is a Research Scientist at the DOE Joint Genome Institute. Her research uses pan-genomics, population genomics, and comparative genomics to explore the functional significance of the variants in plant genomes. Previous and current work has included investigating the abiotic stress responses for the model perennial grass Brachypodium sylvaticum, pan-genomics of Brachypodium species, drought and cold adaption in barley, comparative genomics to explore the variants which could decrease the crop yield, the evolution of gene family related to epigenetics following the whole genome duplications, and comparative transcriptomics in different tissues from different species.
Recent Publications
Related News
Biosciences FY25 LDRD Projects
The projects of 23 Biosciences Area scientists and engineers received funding through the FY25 Laboratory Directed Research and Development (LDRD) program.
Biosciences Area FY24 LDRD Projects
The projects of 21 Biosciences Area scientists and engineers received funding through the FY24 Laboratory Directed Research and Development (LDRD) program.
Divisions
- Science Programs
Secondary Affiliation:
Environmental Genomics and Systems Biology
- Molecular EcoSystems Biology
Research Interests
Bob uses single-cell genomics and metagenomics to explore how microbes influence people and the environment. This includes studying the diversity, physiology and community ecology of microbes sampled from the natural environment.
Recent Publications
Related News
Onsite PhD Student Visit Amps Up Collaborative Spirit
Biosciences Area staff recently hosted 40 PhD students from Wageningen University in the Netherlands over two days at Emery Station East (ESE) and the Integrative Genomics Building (IGB). The group launched their two-week California tour in the Bay Area, stopping by local biotechnology companies and prominent academic research institutions. The contingent visited ESE to tour the facility, make presentations, and discuss potential collaborations. At the IGB, the students attended a day-long symposium that included short talks, tours of several user facilities, and a poster reception.
JGI Demonstrates the Power of One, Amplified
Continuing explorations into a remote hot spring, deep within the British Columbia backcountry, researchers from the University of Calgary and the JGI employed single-cell sequencing to assess the diversity within and between microbial populations. The work, published in The ISME Journal, showed the value of conducting large-scale single cell genomics by collecting nearly 500 single cells from a single low diversity hot spring sediment sample. Their work showed that single cell genomics can add significant value to the other commonly used culture-independent sequencing approaches including amplicon and metagenomic sequencing. Learn more here on the JGI website.
Divisions
Chemical Sciences Division
Secondary Affiliation:
Molecular Biophysics and Integrated Bioimaging
- Bioenergetics
Biography
B.A., Molecular Cell Biology, UC Berkeley, 2009
Ph.D., Chemistry, Caltech, 2014
Postdoctoral researcher, LBNL, 2014-2017 – f-element photospectroscopy
Research Interests
Fundamental & applied dark electrochemistry/photoelectrochemistry
Photophysics of lanthanide downconversion & spectral shifting
Algorithms for Tafel fitting
Inorganic emulation of biophysical systems/processes
Biochemical systems for atmospheric CO2 capture
Programs & Initiatives
Recent Publications
Related News
Biosciences Area FY23 LDRD Projects
The projects of 22 Biosciences Area scientists and engineers received funding through the FY23 Laboratory Directed Research and Development (LDRD) program.
Using Bacteria to Accelerate Carbon Dioxide Capture in Oceans
Peter Agbo, a staff scientist in the Chemical Sciences Division, with a secondary appointment in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division, has proposed a novel method for direct ocean capture of carbon using microbes. Removing CO2 from the oceans will enable them to continue to do their job of absorbing excess CO2 from the atmosphere.
Divisions
- Genomic Technologies
Secondary Affiliation:
Environmental Genomics and Systems Biology
- Biosystems Data Science
Research Interests
My research interests center on systems biology and population genetics, and developing frameworks to draw insight from large-scale ‘omics data. I’m especially interested in using computational tools to understand how communities of all kinds–from collections of functionally specialized plant cells, to co-occurring sets of diverse microbial species, to genotypically heterogenous fruitfly populations–come together as a functional whole, and adapt to changing environments. Currently, my focus is on analyzing data from single cell technologies to profile individual community members, and integrating multi-omic data types to explore community function.
Recent Publications
Related News
JGI, JBEI Partner on Successful RENEW Proposals
The Joint BioEnergy Institute (JBEI) and Joint Genome Institute (JGI) are part of three DOE-funded initiatives under the RENEW program, which supports internships, training, and mentoring to foster diverse talent in the energy workforce.
Divisions
- Genomic Technologies
Secondary Affiliation:
Environmental Genomics and Systems Biology
- Molecular EcoSystems Biology
Research Interests
I work in the field of microbiology and synthetic biology, with a focus on technology development to facilitate strain engineering and microbial functional genomics studies. My background was in molecular genetics, and I use both traditional genetics approaches, and also cutting edge CRISPR-Cas9, Cre-lox recombineering techniques, to genetically engineer microbes from single gene to whole genome levels, in both aerobic and anaerobic organisms. I am passionate about how we could engineering microbes for useful outcomes, especially for new intervention in the prevention and treatment of environmental problems, and human health.
Recent Publications
Related News
Hualan Liu, Microbe Modifier
Liu, a research scientist with the Joint Genome Institute (JGI)’s strain engineering platform, believes in solving real-world problems with synthetic biology. Liu is an expert at manipulating microbial DNA to carry out new functions and helping JGI's user community answer experimental questions.
Building: 91, Room 400D4
Mail Stop: 91R0183
Phone: (925) 296-5832
sjungbluth@lbl.gov
Links
Research Interests
Exploration of Earth’s microbial and viral diversity is a major scientific frontier. Sequencing DNA – the blueprint of life – collected directly from the environment provides a powerful way to access the uncatalogued microbial diversity on our planet. These uncharacterized lifeforms are ripe with novel functions, metabolisms, the potential for biotechnological and medical applications, and will inform us on the evolution of all life.
My previous work has explored the nature and extent of microbial life underground and below the seafloor using tools rooted in molecular biology, biogeochemistry, and microbiology. Now I am working to explore novel microbial diversity in additional environments through the leveraging of ultra-powerful computational resources and true access to massive genome databases (i.e. big data).
Recent Publications

Building: 978, Room 3205
Mail Stop: 978-3200
pengduliu@lbl.gov
https://abpdu.lbl.gov/people/dupeng-liu/
Research Interests
Dr. Liu’s research is dedicated to the use of cutting-edge technologies for the production of renewable fuels and chemicals. His research interests are in catalysis, reaction engineering and process design. In particular, his research harnesses the unconventional solvents, such as supercritical fluids and gas-expanded liquids, as well as advanced membrane technology, e.g., nano filtration to develop resource-efficient process with reduced environmental footprint. At the ABPDU, he plans, leads, and executes process development and piloting related projects in collaboration with R&D sponsors.
Recent Publications
Related News
Biosciences Area Researchers Place at Berkeley Lab Pitch Competition
ABPDU's Asun Oka and Dupeng Liu placed second and third at the event co-hosted by the Lab’s Intellectual Property Office (IPO) and UC Berkeley’s Haas Business School.
Multi-lab Separations Consortium Aims to Decarbonize Biofuels
The Bioprocessing Separations Consortium, originally established in 2016 by the U.S. Department of Energy’s (DOE) Bioenergy Technologies Office (BETO) and led by Argonne National Laboratory, recently received a three year funding renewal to continue advancing separations technologies critical to converting biomass to low-carbon biofuel. The Biosciences Area’s Advanced Biofuels and Bioproducts Process Development Unit (ABPDU) represents Berkeley Lab as a partner in the consortium.
Research Interests
Deepanwita Banerjee, PhD, is currently working on computationally driven host engineering for growth-coupled production of biofuels and bioproducts using the Product Substrate Pairing (PSP) approach. She uses computational strain optimization methods and genome scale metabolic models (GSM) to predict gene targets for engineering Pseudomonas putida. She is also working towards predicting PSP strategies in P. putida for utilization of lignocellulose derived carbon sources for bioenergy and biomanufacturing applications. The goal is to test and learn to better design robust and sustainable microbial hosts that maintain desirable phenotype across industrially relevant scales and conditions.
Deepanwita Banerjee’s research interests include understanding microbial systems for desirable phenotypes using multi-omics integrated systems biology approach. The applications are diverse ranging from improving production of value-added products or assessing microenvironments for a redox imbalance in vivo. She is also interested in understanding the emergent properties of a microbial hosts that are a result of the complex relationship between metabolism and transcriptional regulation. Currently there are digital resources available for model microbes that help inform such relationships but is lacking for non-model microbial systems. Genome-scale network reconstruction of a microbial system requires integration of more than one layer of biological networks (e.g., metabolism, regulation or signaling) as constraints to improve prediction power of such computational models for desirable phenotypes such as growth coupled production or maximum bioconversion of carbon sources.
Programs & Initiatives
Recent Publications
Related News
Speeding up Biomanufacturing with a Turnkey Framework
Researchers from the Joint BioEnergy Institute (JBEI) developed a new framework that reduces the time of developing novel bioproducts. This new workflow, called Product Substrate Pairing (PSP), has already shown great promise for engineering strains that can convert common bacterial food sources into target molecules.
Deepanwita Banerjee, Multi-faceted Modeler
Deepanwita Banerjee learned the value of reusing things at an early age. This perspective continues to influence her life: from her DIY dollhouse project to her research that's recycling genes to build more sustainable products.
Microbe “Rewiring” Technique Promises a Boom in Biomanufacturing
A new approach to modifying microbes’ metabolic processes will speed up production of innovative bio-based fuels, materials, and chemicals Researchers from Lawrence Berkeley National Laboratory (Berkeley Lab) have achieved unprecedented success in modifying a microbe to efficiently produce a compound of interest using a computational model and CRISPR-based gene editing.
Divisions
- Science Programs
Secondary Affiliation:
Environmental Genomics and Systems Biology
- Comparative and Functional Genomics
Biography
Stephen Mondo completed his PhD training at Cornell University in 2013, where he studied host-endosymbiont interactions with Dr. Teresa Pawlowska. After a brief postdoc in the Bogdanove lab at Cornell studying TAL effectors present in fungal endosymbiont he joined the Fungal amd Algal genomics team at the DOE Joint Genome Institute as a Data Scientist. Here, Stephen supports the fungal scientific community through annotation of fungal genomes, comparative genomic analyses for user-driven publications and tool development for improved quality of fungal genome annotations. Stephen also conducts his own independent research in support of DOE mission objectives, primarily in the areas of epigenomics an chromatin regulation. Additionally, he now leads multi-omics data analysis, integration and visualization efforts within the Fungal and Algal genomics program.
Research Interests
Epigenetics, chromatin regulation, fungal comparative genomics, evolutionary & co-evolutionary theory, host-endosymbiont interactions, phylogenomics, early-diverging fungi
Recent Publications
Related News
Biosciences FY25 LDRD Projects
The projects of 23 Biosciences Area scientists and engineers received funding through the FY25 Laboratory Directed Research and Development (LDRD) program.
Biosciences Area FY24 LDRD Projects
The projects of 21 Biosciences Area scientists and engineers received funding through the FY24 Laboratory Directed Research and Development (LDRD) program.
Biography
From the Molecular Foundry page:
Dr. Ajo-Franklin has been a Staff Scientist at the Molecular Foundry since 2007. Before that, she received her Ph.D. in Chemistry from Stanford University with Prof. Steve Boxer and was a post-doctoral fellow with Prof. Pam Silver in the Department of Systems Biology at Harvard Medical School.
Dr. Ajo-Franklin is fascinated by the incredible, diverse functionality of biological molecules and nanoassemblies, and seeks to engineer these complexes and their host organisms to address global challenges in energy and the environment.
Research Interests
Dr. Ajo-Franklin’s research creates engineered bioassemblies–electron nanoconduits–that direct the flow of charge between living cells and non-living materials. Though a combinatorial approach, she has established an unparalleled ability to genetically program the enormously complex biosynthesis of these electron nanoconduits and to quantitatively characterize their function at the biological-inorganic nanointerface. These capabilities have allowed her to discover design principles that govern the emergent processes of nanoconduit assembly and function. In future work, she will expand the combinatorial space through designed genetic variants and adaptive evolution approaches to dissect fundamental controls on charge transfer at this hybrid nanointerface.
ENGINEERING ELECTRONIC COMMUNICATION
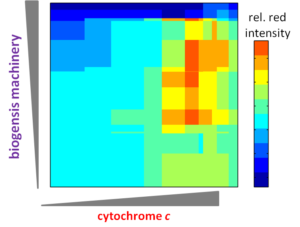
Cellular-electrical connections would enable devices to combine specialties of the living world and the non-living technological world. This new class of smart, self-renewing nanostructured systems has the potential to revolutionize environmental sensing and energy harvesting applications and to open new avenues to program cellular behavior. Building towards this vision, the long-term objectives of the our research are to develop understanding of the principles which govern electron flow across living-non-living interfaces and to use those principles to create electron transfer pathways between individual living microbial cells and non-living electrodes. Our overall strategy is both to explore how naturally-occurring microbes achieve electron transfer to inorganic surfaces and to use genetic and materials surface engineering to create new ‘domesticated’ hybrid cell-electrode systems.
ASSEMBLY & APPLICATIONS OF S-LAYER PROTEINS
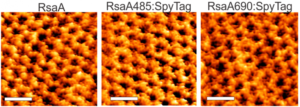
S (`surface’)-layer proteins form crystalline lattices on the outsides of many bacteria and archaea. These nearly ubquitous structures play a number of crucial biological roles: they serve as structural scaffolds, effect selective transport of ions and proteins, serve as templates for mineralization, and protect against phagocytosis. While the lattice structures of many S-layers are known, the dynamical mechanisms through which they form are poorly understood. A molecular-level picture of the assembly of the S-layer protein SbpA from Lysinibacillus sphaericus on lipid bilayers was obtained only recently by using in situ atomic force microscopy. AFM images elucidated the phase transition of amorphous precursors into the crystalline clusters composed of folded tetramers and subsequent growth without disappearance of any crystal clusters. Such a system demonstrates the non-classical crystallization behaviors in the S-layer assembly. Molecular dynamic simulations of the S-layer assembly using a coarse grain model also supported the non-classical nucleation arising from a dense liquid precursor, and subsequent growth of the crystal2. The long-term objectives of our research are to develop predictive understanding of S-layer protein nucleation and growth, so as to advance basic knowledge of the dynamics of coupled nanoscale phase separation and self-assembly and to enable control of the nanoscale structures. Together with the Whitelam, DeYoreo, and Bertozzi groups, we are using a combination of fluorescence microscopy, atomic force microscopy, and computer modeling to reveal the mechanisms underlying S-layer crystallization and to identify strategies for its control.
Recent Publications
Related News
Engineering Living ‘Scaffolds’ for Building Materials
Taking their cue from Nature, Berkeley Lab researchers have engineered living cells to act as a starting point, or scaffold, for the self-assembly of composite materials. The resulting engineered living materials (ELMs) represent a new class of material that may open the door to advanced applications in bioelectronics, biosensing, and smart materials. Leading the effort was Caroline Ajo-Franklin, whose lab is part of the Molecular Foundry, a DOE Office of Science User Facility, and who holds a secondary appointment in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division. A study describing the work was recently published in ACS Synthetic Biology.
Gut Bacteria’s Shocking Secret: They Produce Electricity
UC Berkeley scientists have discovered that a common diarrhea-causing bacterium, Listeria monocytogenes, produces electricity using an entirely different technique from known electrogenic bacteria—and that hundreds of other bacterial species use this same process. The scientists worked Caroline Ajo-Franklin, a staff scientist at the Molecular Foundry who has a secondary appointment in Molecular Biophysics and Integrated Bioimaging, on this research. Read more from the UC Berkeley News Center.
Biosciences and Energy Sciences Areas Host Biomaterials Workshop
On July 16-17, Biosciences, along with the Energy Sciences Area, hosted an internal workshop to discuss and identify paths forward for biomaterials research at Berkeley Lab and in larger contexts–across the national laboratory complex, at universities, and in the private sector– that would enable the creation of new materials with performance-advantaged properties. This workshop continues the program development efforts established through the recent Advanced Biogenic Chemicals and Materials Laboratory-Directed Research and Development initiative established at Berkeley Lab in 2017.