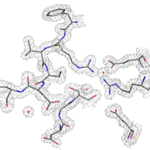
Two recently licensed vaccines against bacterial meningitis contain a bacterial surface protein antigen known as Factor H binding protein (FHbp). The native form of this protein can have low thermal stability, which limits its potential use as an antigen in vaccines. After engineering a more stable Factor H binding protein antigen, scientists from UC San Francisco Benioff Children’s Hospital Oakland determined the structure of the stabilized vaccine with the help of protein crystallography at the Advanced Light Source (ALS) in the Berkeley Center for Structural Biology (Beamline 5.0.1). Read more in the ALS Science Brief.
Raising the Bar for Bacterial Interactions Networks
Scientists working as part of a multi-investigator ENIGMA (Ecosystems & Networks Integrated with Genes and Molecular Assemblies Scientific Focus Area) project have determined that protein-protein interactions occur more frequently among proteins with similar functions. The team of researchers, led by Gareth Butland in the Environmental Genomics & Systems Biology Division, used high throughput functional genomics to study the protein-protein interactome of the model sulfate reducing bacterium Desulfovibrio vulgaris Hildenborough. Their findings critically re-evaluate published bacterial interaction networks and establish benchmarks for high confidence protein interactomes. The manuscript can be found online at Molecular and Cellular Proteomics.
“The Tale of the Bacteria Farmer” with JBEI’s Sarah Richardson
JBEI’s Post-Doctoral Researcher Sarah Richardson (Biological Systems and Engineering Division) teamed up with Team Escamilla at Tumble to record a science podcast for children ages 8 – 12, created to be enjoyed by the entire family. In this podcast episode Richardson explains how she’s trying to convince bacteria to make the things we need, such as biofuels. To listen click here. To learn more about Sarah’s research area and her outreach work click here.
CRISPR/Cas9: Ready for Action
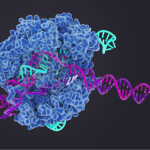
The CRISPR/Cas9 bacterial genomic editing system identifies and cleaves complementary target sequences in foreign DNA. CRISPR (clustered regularly interspaced short palindromic repeats)–associated (Cas) protein Cas9 begins its work by RNA-guided DNA unwinding to form an RNA-DNA hybrid and displacing a DNA strand inside the protein. Upon binding, Cas9 reorganizes into an R-loop complex that is necessary for it to perform its function. A recent article published in Science describes work done to uncover the structural basis of Cas9’s function.
A Key Step Toward Custom-Made Nanoscale Chemical Factories
Scientists have for the first time reengineered a building block of a geometric nanocompartment that occurs naturally in bacteria. The new design provides an entirely new functionality that greatly expands the potential for these compartments to serve as custom-made chemical factories. The work was led by Cheryl Kerfeld, who holds joint appointments with Berkeley Lab’s 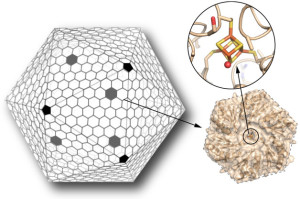 Molecular Biophysics and Integrated Bioimaging Division, UC Berkeley and the MSU-DOE Plant Research Laboratory at Michigan State University. Markus Sutter, a senior research associate in Kerfeld’s group at Berkeley Lab, collected the X-ray diffraction data used in this study in the Berkeley Center for Structural Biology at the Advanced Light Source. Read more at the Berkeley Lab News Center.
Molecular Biophysics and Integrated Bioimaging Division, UC Berkeley and the MSU-DOE Plant Research Laboratory at Michigan State University. Markus Sutter, a senior research associate in Kerfeld’s group at Berkeley Lab, collected the X-ray diffraction data used in this study in the Berkeley Center for Structural Biology at the Advanced Light Source. Read more at the Berkeley Lab News Center.
- « Previous Page
- 1
- …
- 196
- 197
- 198
- 199
- 200
- …
- 213
- Next Page »
Was this page useful?