Many microbial traits of interest to researchers hoping to harness them for applications in biotechnology and human health are controlled by not one, but multiple genes. Understanding how each contributes to a particular function then becomes especially challenging. Yet this knowledge is essential for adapting microbial processes to achieve desired outcomes, such as a novel way of combating antibiotic resistance.
Biosciences Area researchers worked with a team at the National University of Singapore to confirm a new approach that can identify the interactions between genes in bacteria at an unprecedented scale. By simultaneously creating two genetic changes, or mutations, in a single cell, they were able to catalog the effects and ascertain gene function. Senior scientists Adam Arkin and Adam Deutschbauer and research scientist Morgan Price in the Environmental Genomics & Systems Biology (EGSB) Division performed the extensive data analysis associated with the work.
In 2015, a team of Berkeley Lab researchers including Price, Arkin, and Deutschbauer developed a technique known as RB-TnSeq by adding a snippet of DNA that acts as a barcode to mobile genetic elements, called transposons. Researchers could then track the barcoded transposons once they were inserted into bacterial populations and see how well the mutants grew in various conditions.
The new technique, called Dual transposon sequencing (Dual Tn-seq), builds on RB-TnSeq. Researchers constructed pairs of double mutants which, upon analysis, revealed previously unknown genetic interactions and gene functions in Streptococcus pneumoniae, a bacteria that causes respiratory tract infections and more severe diseases like pneumonia and meningitis. This work, the largest single survey of genetic interactions performed on bacteria to date, also identified a new family of enzymes involved in building DNA and RNA. This technique and its application were recently described in Science.
“By looking at pairs of genes instead of just one at a time, we can uncover hidden weaknesses in bacteria that would otherwise go unnoticed,” said Deutschbauer. “This kind of insight not only deepens our understanding of how bacteria work, but also points us toward new strategies for tackling drug-resistant infections.”
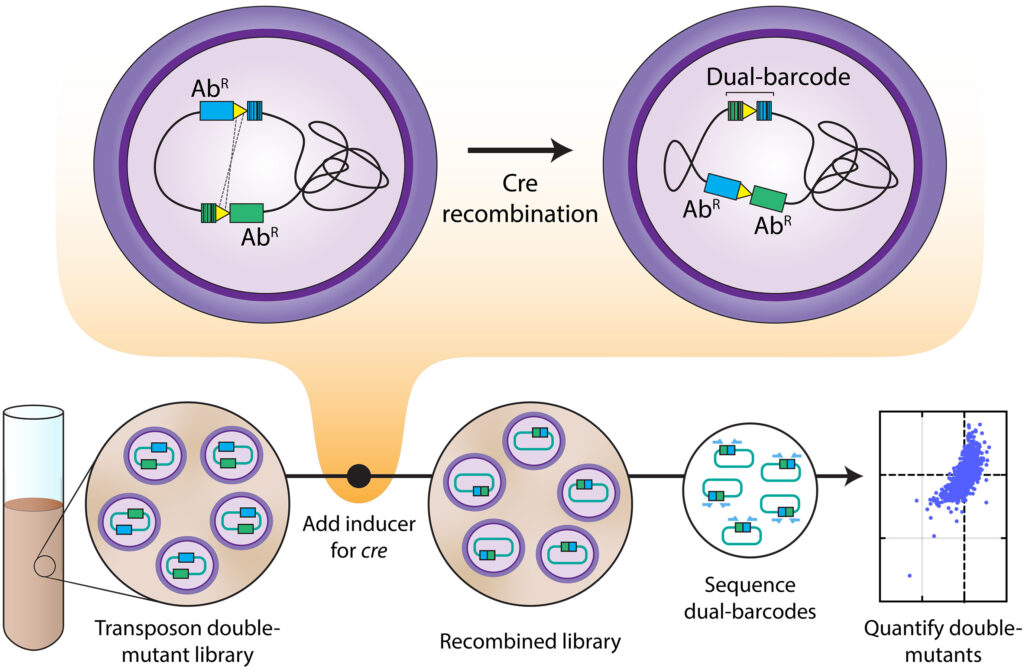
In this work with Dual Tn-seq, the teams generated two random transposon barcodes and used a molecular matchmaker enzyme to move the barcodes close enough to each other so they can be sequenced together. This technique works more quickly and efficiently—over one billion double mutants were created and the teams interrogated about one million gene-to-gene interactions. From these data, the teams uncovered more than 200 previously unknown interactions between genes, including combinations where mutating both genes caused the bacteria to die. They also discovered a new enzyme that produces the building blocks of DNA in disease-causing bacteria, revealing more clues into how many bacteria work.
Reaching beyond the specific discoveries of gene function in S.pneumoniae, this work serves as a benchmark for future studies. “This project is a great example of how a robust technology developed and optimized for environmental bacteria can be modified as a new tool for dissecting gene-gene relationships in a human pathogen, with important implications for novel therapeutic development,” said Deutschbauer.
Read more in the National University of Singapore press release.




