A Berkeley Lab team led by staff scientist Romy Chakraborty explored the impact subtle differences between environments—like those found in varying depths of the same sediment core—have on soil microbe communities. After isolating strains belonging to the genus Arthrobacter, a common soil microbe capable of degrading large, difficult to break down carbon molecules, they found remarkable functional and genomic adaptability correlated with the strain’s environment. Their results, published in International Society for Microbial Ecology Communications, could advance our understanding of global carbon cycling.
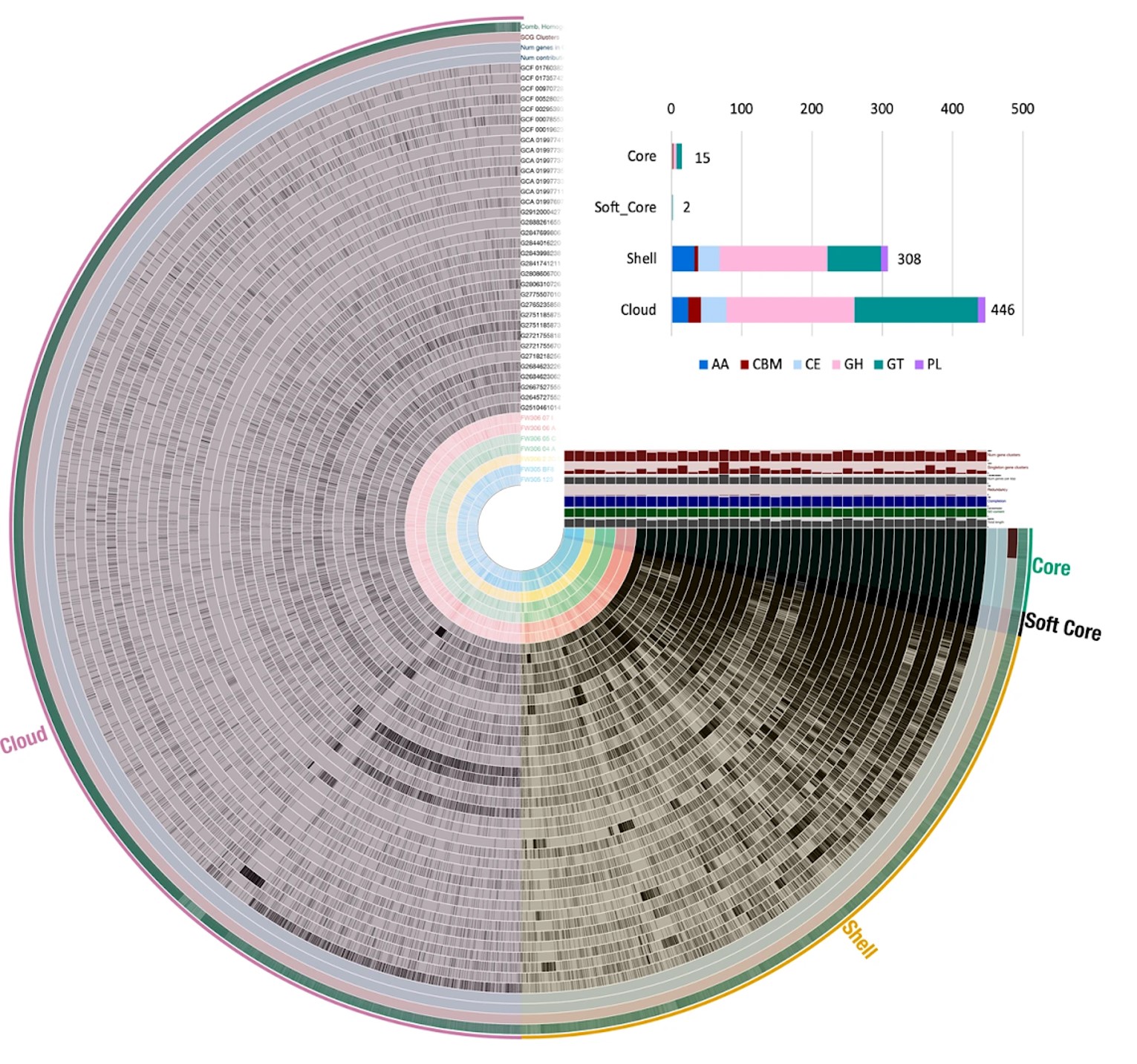
The team analyzed the genotypes and phenotypes of several Arthrobacter strains to correlate cellular functions to their location at varying depths within a single sediment core and in nearby groundwater. Lead author Sara Gushgari-Doyle, a previous postdoctoral researcher in Chakraborty’s group, found that Arthrobacter, as a genus, has remarkable flexibility in altering its suites of carbon degradation genes. This genomic variation was found to be linked to the individual strain’s environment and is the basis for Arthrobacter’s ability to break down a wide variety of complex carbon sources.
With a deeper understanding of how the environment influences a microbe’s genomic potential, and subsequently carbon metabolism function, scientists can begin to tease apart the complex relationships underlying carbon and nutrient cycling and how they might be impacted by a changing climate.
The research team included members of Adam Arkin’s lab at Berkeley Lab, as well as collaborators at the University of Georgia. Chakraborty is also a member of the multi-institutional ENIGMA Science Focus Area led by Arkin and Paul Adams. ENIGMA aims to create a predictive model of microbial communities on ecosystem-level processes.
Learn more on the Earth and Environmental Sciences website.