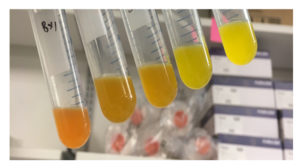
A team led by University of Texas, Austin scientists has developed a more nuanced library approach to tuning gene expression in metabolic pathways. Compared to the traditional way, which leverages an all-or-nothing approach, they can now fine-tune the level of gene expression. In a study, they were able to identify variations of essential genes in metabolic networks that were missed using traditional approaches. The work appeared in the Proceedings of the National Academy of Sciences. Read more on the JGI website.



