Researchers across the world have amassed a wealth of information about the novel coronavirus SARS-CoV-2 and the disease it causes, COVID-19. But this valuable data is stored in different digital libraries, organized in different structures, and written with different jargon. To get the most out of our collective knowledge, someone needs to collect it all in one place.
Under a special project launched in May, a Berkeley Lab–led team of computing and bioinformatics experts is developing a platform that consolidates disparate COVID-19 data sources and uses the unified library to make predictions—about potential drug targets, for example.
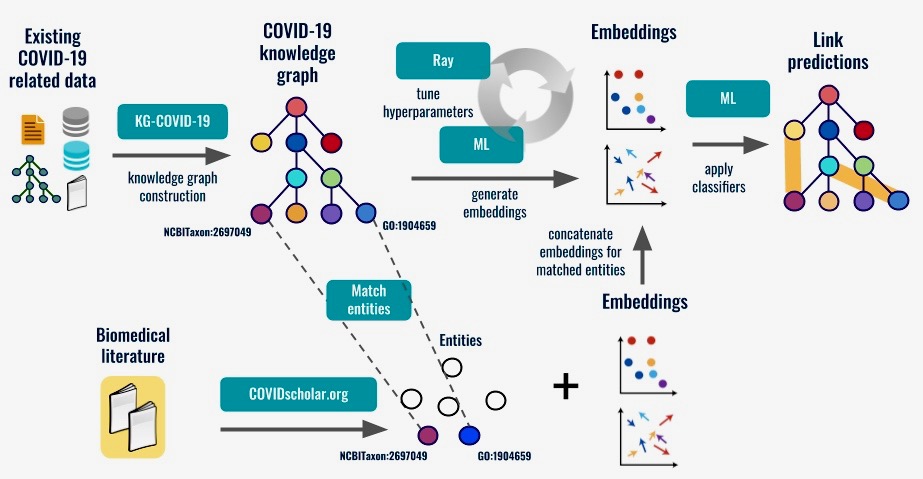
“We’ve built what is called a knowledge graph, where we pull all the various heterogeneous kinds of biological data out there into one location, and organize it according to how the data relate to each other using techniques such as link prediction,” said project principal investigator Chris Mungall, head of the Biosystems Data Science department in Biosciences’ Environmental Genomics and Systems Biology (EGSB) Division.
The knowledge graph is freely available online can be analyzed by open-source software. It currently includes data on approximately 32,000 drugs; 21,000 human and 272 viral proteins plus roughly the same number of genes; and more than 50,000 scientific studies and clinical trials. New relevant information is added as it becomes available.
The next step, which the team is working on now, is to apply machine-learning approaches to the knowledge graph to predict what existing or new drugs could work against COVID-19 based on the properties of those compounds and the viral or human macromolecules they target.
Read more in the Berkeley Lab News Center.