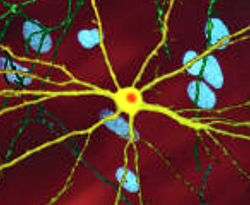
 The mutant form of the Huntington gene, mHTT, which encodes a product that causes the disease, is expressed throughout the brain in affected individuals. Yet neurons in individual regions of the brain are differentially susceptible to its neurotoxic effects. The basis for this puzzling region-specific vulnerability in Huntington disease—which is likewise a feature of Alzheimer and Parkinson neurodegenerative diseases—was hitherto unknown.
The mutant form of the Huntington gene, mHTT, which encodes a product that causes the disease, is expressed throughout the brain in affected individuals. Yet neurons in individual regions of the brain are differentially susceptible to its neurotoxic effects. The basis for this puzzling region-specific vulnerability in Huntington disease—which is likewise a feature of Alzheimer and Parkinson neurodegenerative diseases—was hitherto unknown.
A new study led by Cynthia McMurray, a senior scientist in Molecular Biophysics and Integrated Bioimaging (MBIB), provides evidence that regional differences in neuronal susceptibility to Huntington disease can be attributed to substrate-driven metabolic reprogramming strategies adopted by astrocytes in response to low glucose. By applying a non-invasive, high-sensitivity imaging technique called fluorescence lifetime imaging microscopy (FLIM), the researchers were able to directly measure mitochondrial metabolism in living brain tissues of a mouse model.
The team recently reported their findings in the journal Cell Metabolism.
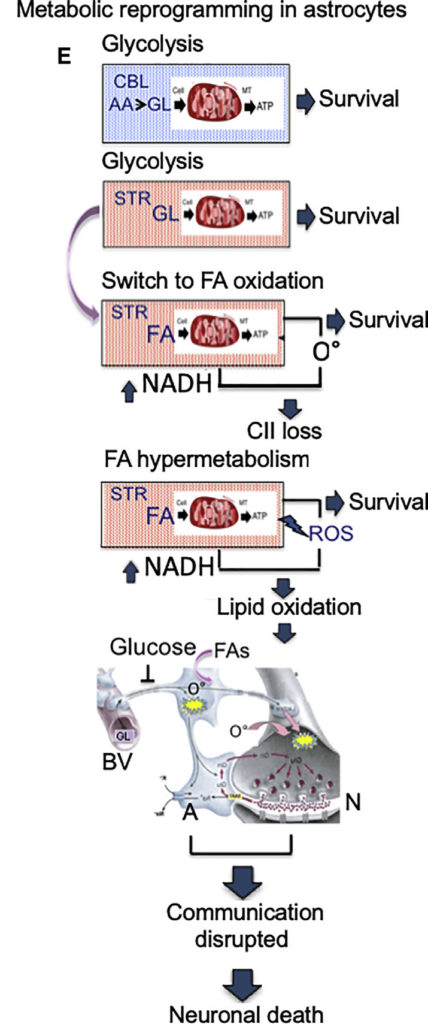
Levels of glucose, the primary fuel of brain cells, are low in the Huntington disease brain. To compensate, the neuron support cells called astrocytes reprogram their mitochondria to use other endogenous energy sources. Each brain region has a distinct pool of available alternative fuels, however, requiring astrocytes to employ different substrate-specific metabolic pathways.
The vulnerable striatum, for example, is enriched in fatty acids so astrocyte mitochondria switch to oxidizing them as an energy source. But this results in the production of reactive oxygen species (ROS) which cause cellular damage. The resistant cerebellum, by contrast, is enriched in amino acid precursors for glucose regeneration, so cells in this area maintain glycolysis and do not sustain ROS-induced damage.
Metabolic reprogramming is potentially a common mechanism by which substrates drive and tailor the survival potential of cells in a heterogeneous environment, noted Aris Polyzos, a research scientist in MBIB and co-first author on the study. “A region-specific metabolic ‘signature’ may provide a general context in which a particular disease protein is able to induce responses which affect one region to a greater extent than another.”
In addition, manipulating the substrate pools may be a viable strategy to control biological organisms in a region-specific manner. “Metabolic reprogramming as an adaptation strategy provides novel pathways for energy production in heterogeneous systems and may have general relevance to artificial photosynthesis, or the response to toxic exposure, disease state, or functional genomic outcomes,” McMurray said.
Additional Biosciences collaborators were: co-first author Do Yup Lee, former affiliate scientist (MBIB); Helen Budworth, affiliate scientist (MBIB); Amy Holt, affiliate scientist (MBIB); Ken Frankel, guest scientist (MBIB); and Kelly Trego, guest scientist (BSE).