Caroline Ajo-Franklin
Visiting Faculty
Professor, Rice University
Biography
From the Molecular Foundry page:
Dr. Ajo-Franklin has been a Staff Scientist at the Molecular Foundry since 2007. Before that, she received her Ph.D. in Chemistry from Stanford University with Prof. Steve Boxer and was a post-doctoral fellow with Prof. Pam Silver in the Department of Systems Biology at Harvard Medical School.
Dr. Ajo-Franklin is fascinated by the incredible, diverse functionality of biological molecules and nanoassemblies, and seeks to engineer these complexes and their host organisms to address global challenges in energy and the environment.
Research Interests
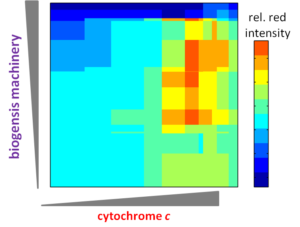
Dr. Ajo-Franklin’s research creates engineered bioassemblies–electron nanoconduits–that direct the flow of charge between living cells and non-living materials. Though a combinatorial approach, she has established an unparalleled ability to genetically program the enormously complex biosynthesis of these electron nanoconduits and to quantitatively characterize their function at the biological-inorganic nanointerface. These capabilities have allowed her to discover design principles that govern the emergent processes of nanoconduit assembly and function. In future work, she will expand the combinatorial space through designed genetic variants and adaptive evolution approaches to dissect fundamental controls on charge transfer at this hybrid nanointerface.
ENGINEERING ELECTRONIC COMMUNICATION
Cellular-electrical connections would enable devices to combine specialties of the living world and the non-living technological world. This new class of smart, self-renewing nanostructured systems has the potential to revolutionize environmental sensing and energy harvesting applications and to open new avenues to program cellular behavior. Building towards this vision, the long-term objectives of the our research are to develop understanding of the principles which govern electron flow across living-non-living interfaces and to use those principles to create electron transfer pathways between individual living microbial cells and non-living electrodes. Our overall strategy is both to explore how naturally-occurring microbes achieve electron transfer to inorganic surfaces and to use genetic and materials surface engineering to create new ‘domesticated’ hybrid cell-electrode systems.
ASSEMBLY & APPLICATIONS OF S-LAYER PROTEINS
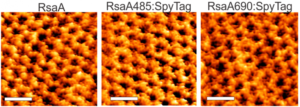
S (`surface’)-layer proteins form crystalline lattices on the outsides of many bacteria and archaea. These nearly ubquitous structures play a number of crucial biological roles: they serve as structural scaffolds, effect selective transport of ions and proteins, serve as templates for mineralization, and protect against phagocytosis. While the lattice structures of many S-layers are known, the dynamical mechanisms through which they form are poorly understood. A molecular-level picture of the assembly of the S-layer protein SbpA from Lysinibacillus sphaericus on lipid bilayers was obtained only recently by using in situ atomic force microscopy. AFM images elucidated the phase transition of amorphous precursors into the crystalline clusters composed of folded tetramers and subsequent growth without disappearance of any crystal clusters. Such a system demonstrates the non-classical crystallization behaviors in the S-layer assembly. Molecular dynamic simulations of the S-layer assembly using a coarse grain model also supported the non-classical nucleation arising from a dense liquid precursor, and subsequent growth of the crystal2. The long-term objectives of our research are to develop predictive understanding of S-layer protein nucleation and growth, so as to advance basic knowledge of the dynamics of coupled nanoscale phase separation and self-assembly and to enable control of the nanoscale structures. Together with the Whitelam, DeYoreo, and Bertozzi groups, we are using a combination of fluorescence microscopy, atomic force microscopy, and computer modeling to reveal the mechanisms underlying S-layer crystallization and to identify strategies for its control.
Recent Publications
Related News
Engineering Living ‘Scaffolds’ for Building Materials
Taking their cue from Nature, Berkeley Lab researchers have engineered living cells to act as a starting point, or scaffold, for the self-assembly of composite materials. The resulting engineered living materials (ELMs) represent a new class of material that may open the door to advanced applications in bioelectronics, biosensing, and smart materials. Leading the effort was Caroline Ajo-Franklin, whose lab is part of the Molecular Foundry, a DOE Office of Science User Facility, and who holds a secondary appointment in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division. A study describing the work was recently published in ACS Synthetic Biology.
Gut Bacteria’s Shocking Secret: They Produce Electricity
UC Berkeley scientists have discovered that a common diarrhea-causing bacterium, Listeria monocytogenes, produces electricity using an entirely different technique from known electrogenic bacteria—and that hundreds of other bacterial species use this same process. The scientists worked Caroline Ajo-Franklin, a staff scientist at the Molecular Foundry who has a secondary appointment in Molecular Biophysics and Integrated Bioimaging, on this research. Read more from the UC Berkeley News Center.
Biosciences and Energy Sciences Areas Host Biomaterials Workshop
On July 16-17, Biosciences, along with the Energy Sciences Area, hosted an internal workshop to discuss and identify paths forward for biomaterials research at Berkeley Lab and in larger contexts–across the national laboratory complex, at universities, and in the private sector– that would enable the creation of new materials with performance-advantaged properties. This workshop continues the program development efforts established through the recent Advanced Biogenic Chemicals and Materials Laboratory-Directed Research and Development initiative established at Berkeley Lab in 2017.